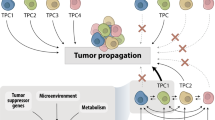
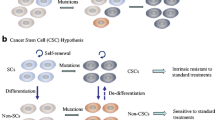
Abstract
Cancer arises from acquired (and sometimes inherited) genetic changes in cells giving rise to a clonal population of neoplastic cells. However, continued growth of a tumor relies on recruitment and subversion of normal stromal elements [2]. In the cell autonomous view of cancer, a tumor is viewed primarily as a genetic disorder whereby a mutation (or series of mutations or other molecular changes) is sufficient to give rise to the malignant state. Thereafter, signaling transduction pathways become deranged, often in ‘oncogene addicted’ states that promote cell growth. In such states, transcriptional programs are co-opted to promote immortalization, resistance to cell senescence and apoptosis, escape of cell cycle checkpoints, promotion of growth of feeding blood vessels (angiogenesis), and ultimately adoption of an invasive and metastatic phenotype. This ‘reductionist’ view of cancer is in keeping with the original six hallmarks of cancer as detailed by Hanahan and Weinberg in 2000 [1]. This viewpoint is a useful construct and effectively synthesizes decades of groundbreaking research to characterize the impact of oncogenic drivers and tumor suppressor genes in cancer cell biology.
The field of cancer research has largely been guided by a reductionist focus on cancer cells and the genes within them—a focus that has produced an extraordinary body of knowledge. Looking forward in time, we believe that important new inroads will come from regarding tumors as complex tissues in which mutant cancer cells have conscripted and subverted normal cell types to serve as active collaborators in their neoplastic agenda. The interactions between the genetically altered malignant cells and these supporting coconspirators will prove critical to understanding cancer pathogenesis and to the development of novel, effective therapies.—Hanahan and Weinberg, Cell 2000 [1]
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Hanahan D, Weinberg RA (2000) The hallmarks of cancer. Cell 100:57–70
Hanahan D, Coussens LM (2012) Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell 21:309–322
Druker BJ, Guilhot F, O'Brien SG, Gathmann I, Kantarjian H, Gattermann N et al (2006) Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N Engl J Med 355:2408–2417
Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144:646–674
Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR et al (2014) Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505:495–501
Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N et al (2009) Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 361:947–957
Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG et al (2010) Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med 363:1693–1703
Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA et al (2010) Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med 363:809–819
Luo J, Solimini NL, Elledge SJ (2009) Principles of cancer therapy: oncogene and non-oncogene addiction. Cell 136:823–837
Perez-Callejo D, Gonzalez-Rincon J, Sanchez A, Provencio M, Sanchez-Beato M (2015) Action and resistance of monoclonal CD20 antibodies therapy in B-cell Non-Hodgkin Lymphomas. Cancer Treat Rev 41:680–689
Cameron F, Sanford M (2014) Ibrutinib: first global approval. Drugs 74:263–271
Markham A (2014) Idelalisib: first global approval. Drugs 74:1701–1707
Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG (2013) Cancer drug resistance: an evolving paradigm. Nat Rev Cancer 13:714–726
Marusyk A, Almendro V, Polyak K (2012) Intra-tumour heterogeneity: a looking glass for cancer? Nat Rev Cancer 12:323–334
Marusyk A, Polyak K (1805) Tumor heterogeneity: causes and consequences. Biochim Biophys Acta 2010:105–117
Michor F, Polyak K (2010) The origins and implications of intratumor heterogeneity. Cancer Prev Res (Phila) 3:1361–1364
Tabassum DP, Polyak K (2015) Tumorigenesis: it takes a village. Nat Rev Cancer 15:473–483
Wood KC (2015) Mapping the pathways of resistance to targeted therapies. Cancer Res 75:4247–4251
Marusyk A, Tabassum DP, Altrock PM, Almendro V, Michor F, Polyak K (2014) Non-cell-autonomous driving of tumour growth supports sub-clonal heterogeneity. Nature 514:54–58
Coppe JP, Patil CK, Rodier F, Sun Y, Munoz DP, Goldstein J et al (2008) Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol 6:2853–2868
Kurtova AV, Xiao J, Mo Q, Pazhanisamy S, Krasnow R, Lerner SP et al (2015) Blocking PGE2-induced tumour repopulation abrogates bladder cancer chemoresistance. Nature 517:209–213
Tissot T, Ujvari B, Solary E, Lassus P, Roche B, Thomas F (1865) Do cell-autonomous and non-cell-autonomous effects drive the structure of tumor ecosystems? Biochim Biophys Acta 2016:147–154
Jouanneau J, Moens G, Bourgeois Y, Poupon MF, Thiery JP (1994) A minority of carcinoma cells producing acidic fibroblast growth factor induces a community effect for tumor progression. Proc Natl Acad Sci U S A 91:286–290
Obenauf AC, Zou Y, Ji AL, Vanharanta S, Shu W, Shi H et al (2015) Therapy-induced tumour secretomes promote resistance and tumour progression. Nature 520:368–372
de Visser KE, Eichten A, Coussens LM (2006) Paradoxical roles of the immune system during cancer development. Nat Rev Cancer 6:24–37
Mueller MM, Fusenig NE (2004) Friends or foes—bipolar effects of the tumour stroma in cancer. Nat Rev Cancer 4:839–849
Masters JR (2000) Human cancer cell lines: fact and fantasy. Nat Rev Mol Cell Biol 1:233–236
Friedman AA, Amzallag A, Pruteanu-Malinici I, Baniya S, Cooper ZA, Piris A et al (2015) Landscape of targeted anti-cancer drug synergies in melanoma identifies a novel BRAF-VEGFR/PDGFR combination treatment. PLoS One 10, e0140310
Friedman AA, Letai A, Fisher DE, Flaherty KT (2015) Precision medicine for cancer with next-generation functional diagnostics. Nat Rev Cancer 15:747–756
Johnson JI, Decker S, Zaharevitz D, Rubinstein LV, Venditti JM, Schepartz S et al (2001) Relationships between drug activity in NCI preclinical in vitro and in vivo models and early clinical trials. Br J Cancer 84:1424–1431
Takimoto CH (2001) Why drugs fail: of mice and men revisited. Clin Cancer Res 7:229–230
Talmadge JE, Singh RK, Fidler IJ, Raz A (2007) Murine models to evaluate novel and conventional therapeutic strategies for cancer. Am J Pathol 170:793–804
Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S et al (2012) The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483:603–607
van Staveren WC, Solis DY, Hebrant A, Detours V, Dumont JE, Maenhaut C (1795) Human cancer cell lines: experimental models for cancer cells in situ? for cancer stem cells? Biochim Biophys Acta 2009:92–103
Lacroix M, Leclercq G (2004) Relevance of breast cancer cell lines as models for breast tumours: an update. Breast Cancer Res Treat 83:249–289
Engelholm SA, Vindelov LL, Spang-Thomsen M, Brunner N, Tommerup N, Nielsen MH et al (1985) Genetic instability of cell lines derived from a single human small cell carcinoma of the lung. Eur J Cancer Clin Oncol 21:815–824
Hausser HJ, Brenner RE (2005) Phenotypic instability of Saos-2 cells in long-term culture. Biochem Biophys Res Commun 333:216–222
Nielsen KV, Madsen MW, Briand P (1994) In vitro karyotype evolution and cytogenetic instability in the non-tumorigenic human breast epithelial cell line HMT-3522. Cancer Genet Cytogenet 78:189–199
Daniel VC, Marchionni L, Hierman JS, Rhodes JT, Devereux WL, Rudin CM et al (2009) A primary xenograft model of small-cell lung cancer reveals irreversible changes in gene expression imposed by culture in vitro. Cancer Res 69:3364–3373
Frese KK, Tuveson DA (2007) Maximizing mouse cancer models. Nat Rev Cancer 7:645–658
Herter-Sprie GS, Kung AL, Wong KK (2013) New cast for a new era: preclinical cancer drug development revisited. J Clin Invest 123:3639–3645
Cheon DJ, Orsulic S (2011) Mouse models of cancer. Annu Rev Pathol 6:95–119
Jin K, Teng L, Shen Y, He K, Xu Z, Li G (2010) Patient-derived human tumour tissue xenografts in immunodeficient mice: a systematic review. Clin Transl Oncol 12:473–480
Monsma DJ, Monks NR, Cherba DM, Dylewski D, Eugster E, Jahn H et al (2012) Genomic characterization of explant tumorgraft models derived from fresh patient tumor tissue. J Transl Med 10:125
Garber K (2009) From human to mouse and back: ‘tumorgraft’ models surge in popularity. J Natl Cancer Inst 101:6–8
Cutz JC, Guan J, Bayani J, Yoshimoto M, Xue H, Sutcliffe M et al (2006) Establishment in severe combined immunodeficiency mice of subrenal capsule xenografts and transplantable tumor lines from a variety of primary human lung cancers: potential models for studying tumor progression-related changes. Clin Cancer Res 12:4043–4054
Tentler JJ, Tan AC, Weekes CD, Jimeno A, Leong S, Pitts TM et al (2012) Patient-derived tumour xenografts as models for oncology drug development. Nat Rev Clin Oncol 9:338–350
Suetsugu A, Katz M, Fleming J, Truty M, Thomas R, Saji S et al (2012) Imageable fluorescent metastasis resulting in transgenic GFP mice orthotopically implanted with human-patient primary pancreatic cancer specimens. Anticancer Res 32:1175–1180
Waters DJ, Janovitz EB, Chan TC (1995) Spontaneous metastasis of PC-3 cells in athymic mice after implantation in orthotopic or ectopic microenvironments. Prostate 26:227–234
Day CP, Carter J, Bonomi C, Hollingshead M, Merlino G (2012) Preclinical therapeutic response of residual metastatic disease is distinct from its primary tumor of origin. Int J Cancer 130:190–199
Voskoglou-Nomikos T, Pater JL, Seymour L (2003) Clinical predictive value of the in vitro cell line, human xenograft, and mouse allograft preclinical cancer models. Clin Cancer Res 9:4227–4239
Chen Z, Cheng K, Walton Z, Wang Y, Ebi H, Shimamura T et al (2012) A murine lung cancer co-clinical trial identifies genetic modifiers of therapeutic response. Nature 483:613–617
Dienstmann R, Jang IS, Bot B, Friend S, Guinney J (2015) Database of genomic biomarkers for cancer drugs and clinical targetability in solid tumors. Cancer Discov 5:118–123
Whiteside TL (2008) The tumor microenvironment and its role in promoting tumor growth. Oncogene 27:5904–5912
Baker BM, Chen CS (2012) Deconstructing the third dimension: how 3D culture microenvironments alter cellular cues. J Cell Sci 125:3015–3024
Villanueva J, Herlyn M (2008) Melanoma and the tumor microenvironment. Curr Oncol Rep 10:439–446
Bissell MJ, Radisky D (2001) Putting tumours in context. Nat Rev Cancer 1:46–54
Tlsty TD, Coussens LM (2006) Tumor stroma and regulation of cancer development. Annu Rev Pathol 1:119–150
Chen F, Zhuang X, Lin L, Yu P, Wang Y, Shi Y et al (2015) New horizons in tumor microenvironment biology: challenges and opportunities. BMC Med 13:45
Mantovani A, Allavena P, Sica A, Balkwill F (2008) Cancer-related inflammation. Nature 454:436–444
Colotta F, Allavena P, Sica A, Garlanda C, Mantovani A (2009) Cancer-related inflammation, the seventh hallmark of cancer: links to genetic instability. Carcinogenesis 30:1073–1081
Fridman WH, Pages F, Sautes-Fridman C, Galon J (2012) The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer 12:298–306
Pardoll DM (2012) The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer 12:252–264
Kamb A (2005) What's wrong with our cancer models? Nat Rev Drug Discov 4:161–165
Sachs N, Clevers H (2014) Organoid cultures for the analysis of cancer phenotypes. Curr Opin Genet Dev 24:68–73
Kenny HA, Lal-Nag M, White EA, Shen M, Chiang CY, Mitra AK et al (2015) Quantitative high throughput screening using a primary human three-dimensional organotypic culture predicts in vivo efficacy. Nat Commun 6:6220
Vaira V, Fedele G, Pyne S, Fasoli E, Zadra G, Bailey D et al (2010) Preclinical model of organotypic culture for pharmacodynamic profiling of human tumors. Proc Natl Acad Sci U S A 107:8352–8356
Baccelli I, Schneeweiss A, Riethdorf S, Stenzinger A, Schillert A, Vogel V et al (2013) Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nat Biotechnol 31:539–544
Hodgkinson CL, Morrow CJ, Li Y, Metcalf RL, Rothwell DG, Trapani F et al (2014) Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer. Nat Med 20:897–903
Yu J, Zhou X, Chang M, Nakaya M, Chang JH, Xiao Y et al (2015) Regulation of T-cell activation and migration by the kinase TBK1 during neuroinflammation. Nat Commun 6:6074
Zhang L, Ridgway LD, Wetzel MD, Ngo J, Yin W, Kumar D et al (2013) The identification and characterization of breast cancer CTCs competent for brain metastasis. Sci Transl Med 5:180ra48
Majumder B, Baraneedharan U, Thiyagarajan S, Radhakrishnan P, Narasimhan H, Dhandapani M et al (2015) Predicting clinical response to anticancer drugs using an ex vivo platform that captures tumour heterogeneity. Nat Commun 6:6169
Nagourney RA, Blitzer JB, Shuman RL, Asciuto TJ, Deo EA, Paulsen M et al (2012) Functional profiling to select chemotherapy in untreated, advanced or metastatic non-small cell lung cancer. Anticancer Res 32:4453–4460
Yu M, Bardia A, Aceto N, Bersani F, Madden MW, Donaldson MC et al (2014) Cancer therapy. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science 345:216–220
Brouzes E, Medkova M, Savenelli N, Marran D, Twardowski M, Hutchison JB et al (2009) Droplet microfluidic technology for single-cell high-throughput screening. Proc Natl Acad Sci U S A 106:14195–14200
Liu X, Ory V, Chapman S, Yuan H, Albanese C, Kallakury B et al (2012) ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. Am J Pathol 180:599–607
Suprynowicz FA, Upadhyay G, Krawczyk E, Kramer SC, Hebert JD, Liu X et al (2012) Conditionally reprogrammed cells represent a stem-like state of adult epithelial cells. Proc Natl Acad Sci U S A 109:20035–20040
Crystal AS, Shaw AT, Sequist LV, Friboulet L, Niederst MJ, Lockerman EL et al (2014) Patient-derived models of acquired resistance can identify effective drug combinations for cancer. Science 346:1480–1486
Fatehullah A, Tan SH, Barker N (2016) Organoids as an in vitro model of human development and disease. Nat Cell Biol 18:246–254
Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S et al (2011) Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology 141:1762–1772
Gao D, Vela I, Sboner A, Iaquinta PJ, Karthaus WR, Gopalan A et al (2014) Organoid cultures derived from patients with advanced prostate cancer. Cell 159:176–187
Boj SF, Hwang CI, Baker LA, Chio II, Engle DD, Corbo V et al (2015) Organoid models of human and mouse ductal pancreatic cancer. Cell 160:324–338
Mengelbier LH, Karlsson J, Lindgren D, Valind A, Lilljebjorn H, Jansson C et al (2015) Intratumoral genome diversity parallels progression and predicts outcome in pediatric cancer. Nat Commun 6:6125
Junttila MR, de Sauvage FJ (2013) Influence of tumour micro-environment heterogeneity on therapeutic response. Nature 501:346–354
Sutherland RM (1988) Cell and environment interactions in tumor microregions: the multicell spheroid model. Science 240:177–184
Hirt C, Papadimitropoulos A, Mele V, Muraro MG, Mengus C, Iezzi G et al (2014) “In vitro” 3D models of tumor-immune system interaction. Adv Drug Deliv Rev 79–80:145–154
Ferrarini M, Steimberg N, Ponzoni M, Belloni D, Berenzi A, Girlanda S et al (2013) Ex-vivo dynamic 3-D culture of human tissues in the RCCS bioreactor allows the study of Multiple Myeloma biology and response to therapy. PLoS One 8, e71613
Ekert JE, Johnson K, Strake B, Pardinas J, Jarantow S, Perkinson R et al (2014) Three-dimensional lung tumor microenvironment modulates therapeutic compound responsiveness in vitro—implication for drug development. PLoS One 9, e92248
Feder-Mengus C, Ghosh S, Reschner A, Martin I, Spagnoli GC (2008) New dimensions in tumor immunology: what does 3D culture reveal? Trends Mol Med 14:333–340
Feder-Mengus C, Ghosh S, Weber WP, Wyler S, Zajac P, Terracciano L et al (2007) Multiple mechanisms underlie defective recognition of melanoma cells cultured in three-dimensional architectures by antigen-specific cytotoxic T lymphocytes. Br J Cancer 96:1072–1082
Palucka AK, Coussens LM (2016) The basis of oncoimmunology. Cell 164:1233–1247
Sharma P, Allison JP (2015) The future of immune checkpoint therapy. Science 348:56–61
Smyth MJ, Ngiow SF, Ribas A, Teng MW (2016) Combination cancer immunotherapies tailored to the tumour microenvironment. Nat Rev Clin Oncol 13:143–158
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Jenkins, R.W. (2017). Introduction to Ex Vivo Cancer Models. In: Aref, A., Barbie, D. (eds) Ex Vivo Engineering of the Tumor Microenvironment. Cancer Drug Discovery and Development. Humana Press, Cham. https://doi.org/10.1007/978-3-319-45397-2_1
Download citation
DOI: https://doi.org/10.1007/978-3-319-45397-2_1
Published:
Publisher Name: Humana Press, Cham
Print ISBN: 978-3-319-45395-8
Online ISBN: 978-3-319-45397-2
eBook Packages: MedicineMedicine (R0)