Abstract
Electronic health records are increasingly useful for conducting secondary observational studies with power that rivals randomized controlled trials.
You have full access to this open access chapter, Download chapter PDF
Similar content being viewed by others
Keywords
FormalPara Take Home Messages-
Electronic health records (EHR) are increasingly useful for conducting secondary observational studies with power that rivals randomized controlled trials.
-
Secondary analysis of EHR data can inform large-scale health systems choices (e.g., pharmacovigilance) or point-of-care clinical decisions (e.g., medication selection).
-
Clinicians, researchers and data scientists will need to navigate numerous challenges facing big data analytics—including systems interoperability, data sharing, and data security—in order to utilize the full potential of EHR and big data-based studies.
1 Introduction
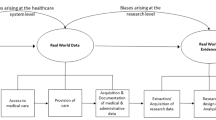
The increased adoption of EHR has created novel opportunities for researchers, including clinicians and data scientists, to access large, enriched patient databases. With these data, investigators are in a position to approach research with statistical power previously unheard of. In this chapter, we present and discuss challenges in the secondary use of EHR data, as well as explore the unique opportunities provided by these data.
2 Challenges in Secondary Analysis of Electronic Health Records Data
Tremendous strides have been made in making pooled health records available to data scientists and clinicians for health research activities, yet still more must be done to harness the full capacity of big data in health care. In all health related fields, the data-holders—i.e., pharmaceutical firms, medical device companies, health systems, and now burgeoning electronic health record vendors—are simultaneously facing pressures to protect their intellectual capital and proprietary platforms, ensure data security, and adhere to privacy guidelines, without hindering research which depends on access to these same databases. Big data success stories are becoming more common, as highlighted below, but the challenges are no less daunting than they were in the past, and perhaps have become even more demanding as the field of data analytics in healthcare takes off.
Data scientists and their clinician partners have to contend with a research culture that is highly competitive—both within academic circles, and among clinical and industrial partners. While little is written about the nature of data secrecy within academic circles, it is a reality that tightening budgets and greater concerns about data security have pushed researchers to use such data as they have on-hand, rather than seek integration of separate databases. Sharing data in a safe and scalable manner is extremely difficult and costly or impossible even within the same institution. With access to more pertinent data restricted or impeded, statistical power and the ability for longitudinal analysis are reduced or lost. None of this is to say researchers have hostile intentions—in fact, many would appreciate the opportunity for greater collaboration in their projects. However, the time, funding, and infrastructure for these efforts are simply deficient. Data is also often segregated into various locales and not consistently stored in similar formats across clinical or research databases. For example, most clinical data is kept in a variety of unstructured formats, making it difficult to query directly via digital algorithms [1]. Within many hospitals, emergency department or outpatient clinical data may exist separately from the hospital and the Intensive Care Unit (ICU) electronic health records, so that access to one does not guarantee access to the other. Images from Radiology and Pathology are typically stored separately in yet other different systems and therefore are not easily linked to outcomes data. The Medical Information Mart for Intensive Care (MIMIC) database described later in this chapter, which contains ICU EHR data from the Beth Israel Deaconess Medical Center (BIDMC), addresses and resolves these artificial divisions, but requires extensive engineering and support staff not afforded to all institutions.
After years of concern about data secrecy, the pharmaceutical industry has recently turned a corner, making detailed trial data available to researchers outside their organizations. GlaxoSmithKline was among the first in 2012 [2], followed by a larger initiative—the Clinical Trial Data Request—to which other large pharmaceutical firms have signed-on [3]. Researchers can apply for access to large-scale information, and integrate datasets for meta-analysis and other systematic reviews. The next frontier will be the release of medical records held at the health system level. The 2009 Health Information Technology for Economic and Clinical Health (HITECH) Act was a boon to the HIT sector [4], but standards for interoperability between record systems continue to lag [5]. The gap has begun to be resolved by government sponsored health information exchanges, as well as the creation of novel research networks [6, 7], but most experts, data scientists, and working clinicians continue to struggle with incomplete data.
Many of the commercial and technical roadblocks alluded to above have their roots in the privacy concerns held by vendors, providers and their patients. Such concerns are not without merit—data breaches of large health systems are becoming distressingly common [8]. Employees of Partners Healthcare in Boston were recently targeted in a “phishing” scheme, unwittingly providing personal information that allowed hackers unauthorized access to patient information [9]; patients of Seton Healthcare in Texas suffered a similar breach just a few months prior [10]. Data breaches aren’t limited to healthcare providers—80 million Anthem enrollees may have suffered loss of their personal information to a cyberattack, the largest of its kind to-date [11]. Not surprisingly in the context of these breaches, healthcare companies have some of the lowest scores of all industries in email security and privacy practices [12]. Such reports highlight the need for prudence amidst exuberance when utilizing pooled electronic health records for big data analytics—such use comes with an ethical responsibility to protect population- and personal-level data from criminal activity and other nefarious ends. For this purpose, federal agencies have convened working groups and public hearings to address gaps in health information security, such as the de-identification of data outside HIPAA-covered entities, and consensus guidelines on what constitutes “harm” from a data breach [13].
Even when issues of data access, integrity, interoperability, security and privacy have been successfully addressed, substantial infrastructure and human capital costs will remain. Though the marginal cost of each additional big data query is small, the upfront cost to host a data center and employ dedicated data scientists can be significant. No figures exist for the creation of a healthcare big data center, and these figures would be variable anyway, depending on the scale and type of data. However, it should not be surprising that commonly cited examples of pooled EHRs with overlaid analytic capabilities—MIMIC (BIDMC), STRIDE (Stanford), the MemorialCare data mart (Memorial Health System, California, $2.2 Billion annual revenue), and the High Value Healthcare Collaborative (hosted by Dartmouth, with 16 other members and funding from the Center for Medicare and Medicaid Services) [14]—come from large, high revenue healthcare systems with regional big-data expertise.
In addition to the above issues, the reliability of studies published using big data methods is of significant concern to experts and physicians. The specific issue is whether these studies are simply amplifications of low-level signals that do not have clinical importance, or are generalizable beyond the database from which they are derived. These are genuine concerns in a medical and academic atmosphere already saturated with innumerable studies of variable quality. Skeptics are concerned that big data analytics will only, “add to the noise,” diverting attention and resources from other venues of scientific inquiry, such as the traditional randomized controlled clinical trial (RCT). While the limitations of RCTs, and the favorable comparison of large observational study results to RCT findings are discussed below, these sentiments nevertheless have merit and must be taken seriously as secondary analysis of EHR data continues to grow. Thought leaders have suggested expounding on the big data principles described above to create open, collaborative learning environments, whereby de-identified data can be shared between researchers—in this manner, data sets can be pooled for greater power, or similar inquiries run on different data sets to see if similar conclusions are reached [15]. The costs for such transparency could be borne by a single institution—much of the cost of creating MIMIC has already been invested, for instance, so the incremental cost of making the data open to other researchers is minimal—or housed within a dedicated collaborative—such as the High Value Healthcare Collaborative funded by its members [16] or PCORnet, funded by the federal government [7]. These collaborative ventures would have transparent governance structures and standards for data access, permitting study validation and continuous peer review of published and unpublished works [15], and mitigating the effects of selection bias and confounding in any single study [17].
As pooled electronic health records achieve even greater scale, data scientists, researchers and other interested parties expect that the costs of hosting, sorting, formatting and analyzing these records are spread among a greater number of stakeholders, reducing the costs of pooled EHR analysis for all involved. New standards for data sharing may have to come into effect for institutions to be truly comfortable with records-sharing, but within institutions and existing research collaboratives, safe practices for data security can be implemented, and greater collaboration encouraged through standardization of data entry and storage. Clear lines of accountability for data access should be drawn, and stores of data made commonly accessible to clarify the extent of information available to any institutional researcher or research group. The era of big data has arrived in healthcare, and only through continuous adaptation and improvement can its full potential be achieved.
3 Opportunities in Secondary Analysis of Electronic Health Records Data
The rising adoption of electronic health records in the U.S. health system has created vast opportunities for clinician scientists, informaticians and other health researchers to conduct queries on large databases of amalgamated clinical information to answer questions both large and small. With troves of data to explore, physicians and scientists are in a position to evaluate questions of clinical efficacy and cost-effectiveness—matters of prime concern in 21st century American health care—with a qualitative and statistical power rarely before realized in medical research. The commercial APACHE Outcomes database, for instance, contains physiologic and laboratory measurements from over 1 million patient records across 105 ICUs since 2010 [18]. The Beth Israel Deaconess Medical Center—a tertiary care hospital with 649 licensed beds including 77 critical care beds—provides an open-access single-center database (MIMIC) encompassing data from over 60,000 ICU stays [19].
Single- and multi-center databases such as those above permit large-scale inquiries without the sometimes untenable expense and difficulty of a randomized clinical trial (RCT), thus answering questions previously untestable in RCTs or prospective cohort studies. This can also be done with increased precision in the evaluation of diagnostics or therapeutics for select sub-populations, and for the detection of adverse events from medications or other interventions with greater expediency, among other advantages [20]. In this chapter, we offer further insight into the utility of secondary analysis of EHR data to investigate relevant clinical questions and provide useful decision support to physicians, allied health providers and patients.
4 Secondary EHR Analyses as Alternatives to Randomized Controlled Clinical Trials
The relative limitations of RCTs to inform real-world clinical decision-making include the following: many treatment comparisons of interest to clinicians have not been addressed by RCTs; when RCTs have been performed and appraised, half of systemic reviews of RCTs report insufficient evidence to support a given medical intervention; and, there are realistic cost and project limitations that prevent RCTs from exploring specific clinical scenarios. The latter include rare conditions, clinically uncommon or disparate events, and a growing list of combinations of recognized patient sub-groups, concurrent conditions (genetic, chronic, acute and healthcare-acquired), and diagnostic and treatment options [20, 21].
Queries on EHR databases to address clinical questions are essentially large, nonrandomized observational studies. Compared to RCTs, they are relatively more efficient and less expensive to perform [22], the majority of the costs having been absorbed by initial system installation and maintenance, and the remainder consisting primarily of research personnel salaries, server or cloud space costs. There is literature to suggest a high degree of correlation between treatment effects reported in nonrandomized studies and randomized clinical trials. Ioannidis et al. [23] found significant correlation (Spearman coefficient of 0.75, p < 0.001) between the treatment effects reported in randomized trials versus nonrandomized studies across 45 diverse topics in general internal medicine, ranging from anticoagulation in myocardial infarction to low-level laser therapy for osteoarthritis. Of particular interest, significant variability in reported treatment outcome “was seen as frequently among the randomized trials as between the randomized and nonrandomized studies,” and they observed that variability was common among both randomized trials and nonrandomized studies [23]. It is worth pointing out that larger treatment effects were more frequently reported in nonrandomized studies than randomized trials (exact p = 0.009) [23]; however, this need not be evidence of publication bias, as relative study size and conservative trial protocol could also cause this finding. Ioannidis et al.’s [24] results are echoed by a more recent Cochrane meta-analysis, which found no significant difference in effect estimates between RCTs and observational studies regardless of the observational study design or heterogeneity.
To further reduce confounding in observational studies, researchers have employed propensity scoring [25], which allows balancing of numerous covariates between treatment groups as well as stratification of samples by propensity score for more nuanced analysis [26]. Kitsios and colleagues matched 18 unique propensity score studies in the ICU setting with at least one RCT evaluating the same clinical question and found a high degree of agreement between their estimates of relative risk and effect size. There was substantial difference in the magnitude of effect sizes in a third of comparisons, reaching statistically significance in one case [27]. Though the RCT remains atop the hierarchy of evidence-based medicine, it is hard to ignore the power of large observational studies that include adequate adjusting for covariates, such as carefully performed studies derived from review of EHRs. The scope of pooled EHR data—whether sixty thousand or one million records—affords insight into small treatment effects that may be under-reported or even missed in underpowered RCTs. Because costs are small compared to RCTs, it is also possible to investigate questions where realistically no study-sponsor will be found. Finally, in the case of databased observational studies, it becomes much more feasible to improve and repeat, or simply repeat, studies as deemed necessary to investigate accuracy, heterogeneity of effects, and new clinical insights.
5 Demonstrating the Power of Secondary EHR Analysis: Examples in Pharmacovigilance and Clinical Care
The safety of pharmaceuticals is of high concern to both patients and clinicians. However, methods for ensuring detection of adverse events post-release are less robust than might be desirable. Pharmaceuticals are often prescribed to a large, diverse patient population that may have not been adequately represented in pre-release clinical trials. In fact, RCT cohorts may deliberately be relatively homogeneous in order to capture the intended effect(s) of a medication without “noise” from co-morbidities that could modulate treatment effects [28]. Humphreys and colleagues (2013) reported that in highly-cited clinical trials, 40 % of identified patients with the condition under consideration were not enrolled, mainly due to restrictive eligibility criteria [29]. Variation in trial design (comparators, endpoints, duration of follow-up) as well as trial size limit their ability to detect low-frequency or long-term side-effects and adverse events [28]. Post-market surveillance reports are imperfectly collected, are not regularly amalgamated, and may not be publically accessible to support clinical-decision making by physicians or inform decision-making by patients.
Queries on pooled EHRs—essentially performing secondary observational studies on large study populations—could compensate for these gaps in pharmacovigilance. Single-center approaches for this and similar questions regarding medication safety in clinical environments are promising. For instance, the highly publicized findings of the Kaiser Study on Vioxx® substantiated prior suspicions of an association between celecoxib and increased risk of serious coronary heart disease [30]. These results were made public in April 2004 after presentation at an international conference; Vioxx® was subsequently voluntarily recalled from the market in September of the same year. Graham and colleagues were able to draw on 2,302,029 person-years of follow-up from the Kaiser Permanente database, to find 8143 cases of coronary heart disease across all NSAIDs under consideration, and subsequently drill-down to the appropriate odds ratios [31].
Using the MIMIC database mentioned above, researchers at the Beth Israel Deaconess Medical Center were able to describe for the first time an increased mortality risk for ICU patients who had been on selective serotonin reuptake inhibitors prior to admission [32]. A more granular analysis revealed that mortality varied by specific SSRI, with higher mortality among patients taking higher-affinity SSRIs (i.e., those with greater serotonin inhibition); on the other hand, mortality could not be explained by common SSRI adverse effects, such as impact on hemodynamic variables [32].
The utility of secondary analysis of EHR data is not limited to the discovery of treatment effects. Lacking published studies to guide their decision to potentially anticoagulate a pediatric lupus patient with multiple risk factors for thrombosis, physicians at Stanford turned to their own EHR-querying platform (the Stanford Translational Research Integrated Database Environment—STRIDE) to create an electronic cohort of pediatric lupus patients to study complications from this illness [33]. In four hours’ time, a single clinician determined that patients with similar lupus complications had a high relative risk of thrombosis, and the decision was made to administer anticoagulation [33].
6 A New Paradigm for Supporting Evidence-Based Practice and Ethical Considerations
Institutional experiences such as those above, combined with evidence supporting the efficacy of observational trials to adequately inform clinical practice, validate the concept of pooled EHRs as large study populations possessing copious amounts of information waiting to be tapped for clinical decision support and patient safety. One can imagine a future clinician requesting a large or small query such as those described above. Such queries might relate to the efficacy of an intervention across a subpopulation, or for a single complicated patient whose circumstances are not satisfactorily captured in any published trial. Perhaps this is sufficient for the clinician to recommend a new clinical practice; or maybe they will design a pragmatic observational study for more nuance—evaluating dose-responsiveness, or adverse effect profiles across subpopulations. As clinical decisions are made and the patient’s course of care shaped, this intervention and outcomes information is entered into the electronic health record, effectively creating a feedback loop for future inquiries [34].
Of course, the advantages of secondary analysis of electronic health records must always be balanced with ethical considerations. Unlike traditional RCTs, there is no explicit consent process for the use of demographic, clinical and other potentially sensitive data captured in the EHR. Sufficiently specific queries could yield very narrow results—theoretically specific enough to re-identify an individual patient. For instance, an inquiry on patients with a rare disease, within a certain age bracket, and admitted within a limited timeframe, could include someone who may be known to the wider community. Such an extreme example highlights the need for compliance with federal privacy laws as well as ensuring high institutional standards of data security such as secured servers, limited access, firewalls from the internet, and other data safety methods.
Going further, data scientists should consider additional measures intentionally designed to protect patient anonymity, e.g. date shifting as implemented in the MIMIC database (see Sect. 5.1, Chap. 5). In situations where queries might potentially re-identify patients, such as in the investigation of rare diseases, or in the course of a contagious outbreak, researchers and institutional research boards should seek accommodation with this relatively small subset of potentially affected patients and their advocacy groups, to ensure their comfort with secondary analyses. Disclosure of research intent and methods by those seeking data access might be required, and a patient option to embargo one’s own data should be offered.
It is incumbent on researchers and data scientists to explain the benefits of participation in a secondary analysis to patients and patient groups. Such sharing allows the medical system to create a clinical database of sufficient magnitude and quality to benefit individual- and groups of patients, in real-time or in the future. Also, passive clinical data collection allows the patient to contribute, at relatively very low risk and no personal cost, to the ongoing and future care of others. We believe that people are fundamentally sufficiently altruistic to consider contributions their data to research, provided the potential risks of data usage are small and well-described.
Ultimately, secondary analysis of EHR will only succeed if patients, regulators, and other interested parties are assured and reassured that their health data will be kept safe, and processes for its use are made transparent to ensure beneficence for all.
References
Riskin D (2012) Big data: opportunity and challenge. HealthcareITNews, 12 June 2012. URL: http://www.healthcareitnews.com/news/big-data-opportunity-and-challenge
Harrison C (2012) GlaxoSmithKline opens the door on clinical data sharing. Nat Rev Drug Discov 11(12):891–892. doi:10.1038/nrd3907 [Medline: 23197021]
Clinical Trial Data Request. URL: https://clinicalstudydatarequest.com/. Accessed 11 Aug 2015. [WebCite Cache ID 6TFyjeT7t]
Adler-Milstein J, Jha AK (2012) Sharing clinical data electronically: a critical challenge for fixing the health care system. JAMA 307(16):1695–1696
Verdon DR (2014) ONC’s plan to solve the EHR interoperability puzzle: an exclusive interview with National Coordinator for Health IT Karen B. DeSalvo. Med Econ. URL: http://medicaleconomics.modernmedicine.com/medical-economics/news/onc-s-plan-solve-ehr-interoperability-puzzle?page=full
Green M (2015) 10 things to know about health information exchanges. Becker’s Health IT CIO Rev. URL: http://www.beckershospitalreview.com/healthcare-information-technology/10-things-to-know-about-health-information-exchanges.html
PCORnet. URL: http://www.pcornet.org/. Accessed 11 Aug 2015
Dvorak K (2015) Big data’s biggest healthcare challenge: making sense of it all. FierceHealthIT, 4 May 2015. URL: http://www.fiercehealthit.com/story/big-datas-biggest-healthcare-challenge-making-sense-it-all/2015-05-04
Bartlett J (2015) Partners healthcare reports data breach. Boston Bus J. URL: http://www.bizjournals.com/boston/blog/health-care/2015/04/partners-healthcare-reports-potential-data-breach.html
Dvorak K (2015) Phishing attack compromises info of 39 K at Seton healthcare family. FierceHealthIT, 28 April 2015. URL: http://www.fiercehealthit.com/story/phishing-attack-compromises-info-39k-seton-healthcare-family/2015-04-28
Bowman D (2015) Anthem hack compromises info for 80 million customers. FierceHealthPayer, 5 February 2015. URL: http://www.fiercehealthpayer.com/story/anthem-hack-compromises-info-80-million-customers/2015-02-05
Dvorak K (2015) Healthcare industry ‘behind by a country mile’ in email security. FierceHealthIT, 20 February 2015. URL: http://www.fiercehealthit.com/story/healthcare-industry-behind-country-mile-email-security/2015-02-20
White house seeks to leverage health big data, safeguard privacy. HealthData Manage. URL: http://www.healthdatamanagement.com/news/White-House-Seeks-to-Leverage-Health-Big-Data-Safeguard-Privacy-50829-1.html
How big data impacts healthcare. Harv Bus Rev. URL: https://hbr.org/resources/pdfs/comm/sap/18826_HBR_SAP_Healthcare_Aug_2014.pdf. Accessed 11 Aug 2015
Moseley ET, Hsu DJ, Stone DJ, Celi LA (2014) Beyond open big data: addressing unreliable research. J Med Internet Res 16(11):e259
High value healthcare collaborative. URL: http://highvaluehealthcare.org/. Accessed 14 Aug 2015
Badawi O, Brennan T, Celi LA et al (2014) Making big data useful for health care: a summary of the inaugural mit critical data conference. JMIR Med Inform 2(2):e22
APACHE Outcomes. Available at: https://www.cerner.com/Solutions/Hospitals_and_Health_Systems/Critical_Care/APACHE_Outcomes/. Accessed Nov 2014
Saeed M, Villarroel M, Reisner AT et al (2011) Multiparameter intelligent monitoring in intensive care II (MIMIC-II): a public-access intensive care unit database. Crit Care Med 39:952
Ghassemi M, Celi LA, Stone DJ (2015) State of the art review: the data revolution in critical care. Crit Care 19:118
Mills EJ, Thorlund K, Ioannidis J (2013) Demystifying trial networks and network meta-analysis. BMJ 346:f2914
Angus DC (2007) Caring for the critically ill patient: challenges and opportunities. JAMA 298:456–458
Ioannidis JPA, Haidich A-B, Pappa M et al (2001) Comparision of evidence of treatment effects in randomized and nonrandomized studies. JAMA 286:7
Anglemyer A, Horvath HT, Bero L (2014) Healthcare outcomes assess with observational study designs compared with those assessed in randomized trials. Cochrane Database Syst Rev 29:4
Gayat E, Pirracchio R, Resche-Rigon M et al (2010) Propensity scores in intensive care and anaesthesiology literature: a systematic review. Intensive Care Med 36:1993–2003
Glynn RJ, Schneeweiss S, Stürmer T (2006) Indications for propensity scores and review of their use in pharmacoepidemiology. Basic Clin Pharmacol Toxicol 98:253–259
Kitsios GD, Dahabreh IJ, Callahan S et al (2015) Can we trust observational studies using propensity scores in the critical care literature? A systematic comparison with randomized clinical trials. Crit Care Med (Epub ahead of print)
Celi LA, Moseley E, Moses C et al (2014) from pharmacovigilance to clinical care optimization. Big Data 2(3):134–141
Humphreys K, Maisel NC, Blodgett JC et al (2013) Extent and reporting of patient nonenrollment in influential randomized clinical trials, 2001 to 2010. JAMA Intern Med 173:1029–1031
Vioxx and Drug Safety. Statement of Sandra Kweder M.D. (Deputy Director, Office of New Drugs, US FDA) before the Senate Committee on Finance. Available at: http://www.fda.gov/NewsEvents/Testimony/ucm113235.htm. Accessed July 2015
Graham DJ, Campen D, Hui R et al (2005) Risk of acute myocardial infarction and sudden cardiac death in patients treated with cyclo-oxygenase 2 selective and non-selective non-steroidal anti-inflammatory drugs: nested case-control study. Lancet 365(9458):475–481
Ghassemi M, Marshall J, Singh N et al (2014) Leveraging a critical care database: selective serotonin reuptake inhibition use prior to ICU admission is associated with increased hospital mortality. Chest 145(4):1–8
Frankovich J, Longhurst CA, Sutherland SM (2011) Evidence-based medicine in the EMR era. New Engl J Med 365:19
Celi LA, Zimolzak AJ, Stone DJ (2014) Dynamic clinical data mining: search engine-based decision support. JMIR Med Inform 2(1):e13
Author information
Authors and Affiliations
Rights and permissions
Open Access This chapter is distributed under the terms of the Creative Commons Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/), which permits any noncommercial use, duplication, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, a link is provided to the Creative Commons license and any changes made are indicated.
The images or other third party material in this chapter are included in the work’s Creative Commons license, unless indicated otherwise in the credit line; if such material is not included in the work’s Creative Commons license and the respective action is not permitted by statutory regulation, users will need to obtain permission from the license holder to duplicate, adapt or reproduce the material.
Copyright information
© 2016 The Author(s)
About this chapter
Cite this chapter
Nair, S., Hsu, D., Celi, L.A. (2016). Challenges and Opportunities in Secondary Analyses of Electronic Health Record Data. In: Secondary Analysis of Electronic Health Records. Springer, Cham. https://doi.org/10.1007/978-3-319-43742-2_3
Download citation
DOI: https://doi.org/10.1007/978-3-319-43742-2_3
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-43740-8
Online ISBN: 978-3-319-43742-2
eBook Packages: MedicineMedicine (R0)