Abstract
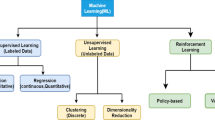
There is a variety of patterns we desire to learn from radiation oncologic data. The previous chapters described how these various learning objectives can commonly be formulated in theoretical nomenclatures. This chapter introduces different machine learning algorithms that could cater to readers’ specific learning goals. We intend to provide conceptual outlines of some of the widely used algorithms with minimal mathematical conundrum and examples drawn from the radiotherapy literature. In this chapter we classify the algorithms into three types, based on the availability of information: unsupervised, supervised, and reinforcement learning. The methods illustrated in this chapter include principal component analysis and clustering (unsupervised), logistic regression, neural network, support vector machine, decision tree, Bayesian networks, and naive Bayes (supervised) in addition to reinforcement learning.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Arthur D, Vassilvitskii S. K-means++: the advantages of careful seeding. In: Proceedings of the eighteenth annual ACM-SIAM symposium on discrete algorithms, SODA’07. Society for Industrial and Applied Mathematics; Philadelphia:2007. p. 1027–35.
Bischof H, Leonardis A, Selb A. Minimum description length principle for robust vector quantisation. Pattern Anal Appl. 1999;2(1):59–72.
Blanco AI, Chao KSC, El Naqa I, Franklin GE, Zakarian K, Vicic M, Deasy JO. Dose-volume modeling of salivary function in patients with head-and-neck cancer receiving radiotherapy. Int J Radiat Oncol Biol Phys. 2005;62(4):1055–69.
Bradley J, Deasy JO, Bentzen S, El-Naqa I. Dosimetric correlates for acute esophagitis in patients treated with radiotherapy for lung carcinoma. Int J Radiat Oncol Biol Phys. 2004;58(4):1106–13.
Bradley JD, Hope A, El Naqa I, Apte A, Lindsay PE, Bosch W, Matthews J, Sause W, Graham MV, Deasy JO. A nomogram to predict radiation pneumonitis, derived from a combined analysis of rtog 9311 and institutional data. Int J Radiat Oncol Biol Phys. 2007;69(4):985–92.
Breiman L. Random forests. Mach Learn. 2001;45(1):5–32.
Chen S, Zhou S, Yin F-F, Marks LB, Das SK. Investigation of the support vector machine algorithm to predict lung radiation-induced pneumonitis. Med Phys. 2007;34(10):3808–14.
Chen S, Zhou S, Yin FF, Marks LB, Das SK. Using patient data similarities to predict radiation pneumonitis via a self-organizing map. Phys Med Biol. 2008;53(1):203.
Chow C, Liu C. Approximating discrete probability distributions with dependence trees. IEEE Trans Inf Theor. 2006;14(3):462–7.
Cooper GF, Herskovits E. A Bayesian method for the induction of probabilistic networks from data. Mach Learn. 1992;9(4):309–47.
Das SK, Zhou S, Zhang J, Yin F-F, Dewhirst MW, Marks LB. Predicting lung radiotherapy-induced pneumonitis using a model combining parametric Lyman probit with nonparametric decision trees. Int J Radiat Oncol Biol Phys. 2007;68(4):1212–21.
Dehing-Oberije C, Yu S, Ruysscher DD, Meersschout S, Beek KV, Lievens Y, Meerbeeck JV, Neve WD, Rao B, van der Weide H, Lambin P. Development and external validation of prognostic model for 2-year survival of non small cell lung cancer patients treated with chemoradiotherapy. Int J Radiat Oncol Biol Phys. 2009;74(2):355–62.
Delaney G, Barton M, Jacob S. Estimation of an optimal radiotherapy utilization rate for melanoma. Cancer. 2004;100(6):1293–301.
El Naqa I, Bradley J, Blanco AI, Lindsay PE, Vicic M, Hope A, Deasy JO. Multivariable modeling of radiotherapy outcomes, including dose-volume and clinical factors. Int J Radiat Oncol Biol Phys. 2006;64(4):1275–86.
El Naqa I, Bradley J, Deasy J. Machine learning methods for radiobiological outcome modeling. In: Mehta M, Paliwal B, Bentzen S, editors. Physical, chemical, and biological targeting in radiation oncology. Madison: Medical Physics Pub.; 2005. p. 150–9.
El Naqa I, Bradley JD, PE L, Hope AJ, Deasy JO. Predicting radiotherapy outcomes using statistical learning techniques. Phys Med Biol. 2009;54(18):S9.
Freund Y, Schapire RE. A brief introduction to boosting. In: Proceedings of the sixteenth international joint conference on artificial intelligence. San Francisco: Morgan Kaufmann; 1999. p. 1401–6.
Friedman JH. On bias, variance, 0/1ñloss, and the curse-of-dimensionality. Data Min Knowl Discov. 1997;1(1):55–77.
Hastie T, Tibshirani R, Friedman J. The elements of statistical learning: data mining, inference, and prediction. New York: Springer; 2009.
Haykin S. Neural networks: a comprehensive foundation. Upper Saddle River: Prentice Hall PTR; 1998.
Hope AJ, Lindsay PE, Naqa IE, Alaly JR, Vicic M, Bradley JD, Deasy JO. Modeling radiation pneumonitis risk with clinical, dosimetric, and spatial parameters. Int J Radiat Oncol Biol Phys. 2006;65(1):112–24.
Hosmer D, Lemeshow S. Applied logistic regression. New York: John Wiley; 2000.
Huang EX, Bradley JD, El Naqa I, Hope AJ, Lindsay PE, Bosch WR, Matthews JW, Sause WT, Graham MV, Deasy JO. Modeling the risk of radiation-induced acute esophagitis for combined Washington University and rtog trial 93-11 lung cancer patients. Int J Radiat Oncol Biol Phys. 2012;82(5):1674–9.
Huang EX, Hope AJ, Lindsay PE, Trovo M, El Naqa I, Deasy JO, Bradley JD. Heart irradiation as a risk factor for radiation pneumonitis. Acta Oncol. 2011;50(1):51–60.
Jain AK, Murty MN, Flynn PJ. Data clustering: a review. ACM Comput Surv. 1999;31(3):264–323.
Jayasurya K, Fung G, Yu S, Dehing-Oberije C, De Ruysscher D, Hope A, De Neve W, Lievens Y, Lambin P, Dekker ALAJ. Comparison of Bayesian network and support vector machine models for two-year survival prediction in lung cancer patients treated with radiotherapy. Med Phys. 2010;37(4):1401–7.
Kazmierska J, Malicki J. Application of the nave Bayesian classifier to optimize treatment decisions. Radiother Oncol. 2008;86(2):211–6.
Kim M, Ghate A, Phillips MH. A Markov decision process approach to temporal modulation of dose fractions in radiation therapy planning. Phys Med Biol. 2009;54(14):4455.
Klement R, Allgauer M, Appold S, Dieckmann K, Ernst I, Ganswindt U, Holy R, Nestle U, Nevinny-Stickel M, Semrau S, Sterzing F, Wittig A, Andratschke N, Guckenberger M. Support vector machine-based prediction of local tumor control after stereotactic body radiation therapy for early-stage non-small cell lung cancer. Int J Radiat Oncol Biol Phys. 2014;88(3):732–8.
Kohonen T. The self-organizing map. Proc IEEE. 1990;78(9):1464–80.
Koller D, Friedman N. Probabilistic graphical models: principles and techniques – adaptive computation and machine learning. Cambridge: The MIT Press; 2009.
Kulkarni P. Reinforcement and systemic machine learning for decision making. Hoboken: Wiley-IEEE Press; 2012.
Madigan D, York J, Allard D. Bayesian graphical models for discrete data. Int Stat Rev. 1995;63(2):215–32.
Marks LB. Dosimetric predictors of radiation-induced lung injury. Int J Radiat Oncol Biol Phys. 2002;54(2):313–6.
Mitchell TM. Machine learning. 1st ed. New York: McGraw-Hill, Inc.; 1997.
Oh JH, Craft JM, Townsend R, Deasy JO, Bradley JD, El Naqa I. A bioinformatics approach for biomarker identification in radiation-induced lung inflammation from limited proteomics data. J Proteome Res. 2011;10(3):1406–15.
Pella A, Cambria R, Riboldi M, Jereczek-Fossa BA, Fodor C, Zerini D, Torshabi AE, Cattani F, Garibaldi C, Pedroli G, Baroni G, Orecchia R. Use of machine learning methods for prediction of acute toxicity in organs at risk following prostate radiotherapy. Med Phys. 2011;38(6):2859–67.
Pelleg D, Moore A. X-means: extending k-means with efficient estimation of the number of clusters. In: Proceedings of the 17th international conference on machine learning. San Francisco: Morgan Kaufmann; 2000. p. 727–34.
Quinlan JR. Induction of decision trees. Mach Learn. 1986;1:81–106.
Quinlan JR. Simplifying decision trees. Int J Man Mach Stud. 1987;27(3):221–34.
Ripley BD. Pattern recognition and neural networks. Cambridge/New York: Cambridge University Press; 1996.
Scholkopf A, Smola J, Muller KR. Kernel principal component analysis. Cambridge: MIT Press; 1999. p. 327–52.
Smith WP, Doctor J, Meyer J, Kalet IJ, Phillips MH. A decision aid for intensity-modulated radiation-therapy plan selection in prostate cancer based on a prognostic Bayesian network and a Markov model. Artif Intell Med. 2009;46(2):119–30.
Specht DF. A general regression neural network. IEEE Trans Neural Netw. 1991;2(6):568–76.
Spencer SJ, Bonnin DA, Deasy JO, Bradley JD, El Naqa I. Bioinformatics methods for learning radiation-induced lung inflammation from heterogeneous retrospective and prospective data. J Biomed Biotechnol. 2009(2009), 892863. doi:10.1155/2009/892863.
Su M, Miften M, Whiddon C, Sun X, Light K, Marks L. An artificial neural network for predicting the incidence of radiation pneumonitis. Med Phys. 2005;32(2):318–25.
Sutton RS, Barto AG. Introduction to reinforcement learning. Cambridge: MIT Press; 1998.
Svensson JP, Stalpers LJA, Lange REEE, Franken NAP, Haveman J, Klein B, Turesson I, Vrieling H, Giphart-Gassler M. Analysis of gene expression using gene sets discriminates cancer patients with and without late radiation toxicity. PLoS Med. 2006;3(10):e422.
Tucker SL, Cheung R, Dong L, Liu HH, Thames HD, Huang EH, Kuban D, Mohan R. Dose-volume response analyses of late rectal bleeding after radiotherapy for prostate cancer. Int J Radiat Oncol Biol Phys. 2004;59(2):353–65.
Vapnik V. Statistical learning theory. New York: Wiley; 1998.
Vittinghoff E, Glidden D, Shiboski S, McCulloch C. Regression methods in biostatistics: linear, logistic, survival, and repeated measures models. New York: Springer; 2006.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Lee, S., El Naqa, I. (2015). Machine Learning Methodology. In: El Naqa, I., Li, R., Murphy, M. (eds) Machine Learning in Radiation Oncology. Springer, Cham. https://doi.org/10.1007/978-3-319-18305-3_3
Download citation
DOI: https://doi.org/10.1007/978-3-319-18305-3_3
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-18304-6
Online ISBN: 978-3-319-18305-3
eBook Packages: MedicineMedicine (R0)