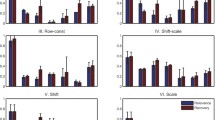
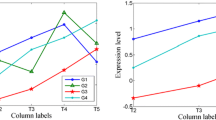
Abstract
Nowadays, biclustering problem is still an intractable problem. But in time series expression data, the clusters can be limited those with contiguous columns. This restriction makes biclustering problem to be tractable problem. However existing contiguous column biclustering algorithm can only find the biclusters which have the same value for each column in biclusters without error tolerance. This characteristic leads the algorithm to overlook some patterns in its clustering process. We propose a suffix tree based algorithm that allows biclusters to have inconsistencies in at most k contiguous column. This can reveals previously undiscoverable biclusters. Our algorithm still has tractable run time with this additional feature.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
McLachlan, G.J., Do, K., Ambroise, C.: Analysing Microarray Gene Expression Data. John Wiley & Sons, New Jersey (2004)
Cheng, Y., Church, G.M.: Biclustering of Expression Data. In: Proc. 8th Int’l Conf. Intelligent Systems for Molecular Biology (ISMB 2000), pp. 93–103. ACM Press, New York (2000)
Madeira, S.C., Oliveira, A.L.: Biclustering Algorithms for Biological Data Analsis: a survey. IEEE/ACM Trans. Computational Biology and Bioinformatics 1(1), 24–45 (2004)
Van Mechelen, I., Bock, H.H., De Boeck, P.: Two-Mode Clustering Methods: A Structured Overview. Statistical Methods in Medical Research 13(5), 979–981 (2004)
Ben-Dor, A., Chor, B., Karp, R., Yakhini, Z.: Discovering Local Structure in Gene Expression Data: The Order-Preserving Submatrix Problem. In: Proc. 6th Int’l Conf. Computational Biology (RECOMB 2002), pp. 49–57 (2002)
Ji, L., Tan, K.: Identifying Time-Lagged Gene Clusters Using Gene Expression Data. Bioinformatics 21(4), 509–516 (2005)
Koyuturk, M., Szpankowski, W., Grama, A.: Biclustering Gene-Feature Matrices for Statistically Significant Dense Patterns. In: Proc. 8th Int’l Conf. Research in Computational Molecular Biology (RECOMB 2004), pp. 480–484 (2004)
Liu, J., Wang, W., Yang, J.: Biclustering in Gene Expression Data by Tendency. In: Proc. 3rd Int’l IEEE CS Computational Systems Bioinformatics Conf. (CSB 2004), pp. 182–193 (2004)
Liu, J., Wang, W., Yang, J.: A Framework for Ontology-Driven Subspace Clustering. In: Proc. ACM SIGKDD 2004, pp. 623–628 (2004)
Liu, J., Wang, W., Yang, J.: Gene Ontology Friendly Biclustering of Expression Profiles. In: Proc. 3rd IEEE CS Computational Systems Bioinformatics Conf. (CSB 2004), pp. 436–447 (2004)
Liu, J., Wang, W., Yang, J.: Mining Sequential Patterns from Large Data Sets. Advances in Database Systems, vol. 18. Kluwer Academic Publishers (2005)
Lonardi, S., Szpankowski, W., Yang, Q.: Finding Biclusters by Random Projections. In: Sahinalp, S.C., Muthukrishnan, S.M., Dogrusoz, U. (eds.) CPM 2004. LNCS, vol. 3109, pp. 102–116. Springer, Heidelberg (2004)
Madeira, S.C., Oliveira, A.L.: A Linear Time Algorithm for Biclustering Time Series Expression Data. In: Casadio, R., Myers, G. (eds.) WABI 2005. LNCS (LNBI), vol. 3692, pp. 39–52. Springer, Heidelberg (2005)
Murali, T.M., Kasif, S.: Extracting Conserved Gene Expression Motifs from Gene Expression Data. In: Proc. 8th Pacific Symp. Biocomputing (PSB 2003), vol. 8, pp. 77–88 (2003)
Prelic, A., Bleuler, S., Zimmermann, P., Wille, A., Bühlmann, P., Gruissem, W., Hennig, L., Thiele, L., Zitzler, E.: A Systematic Comparison and Evaluation of Biclustering Methods for Gene Expression Data. Bioinformatics 22(10), 1282–1283 (2006)
Sheng, Q., Moreau, Y., De Moor, B.: Biclustering Microarray Data by Gibbs Sampling. Bioinformatics 19(2), 196–205 (2003)
Tanay, A., Sharan, R., Shamir, R.: Discovering Statiscally Significant Biclusters in Gene Expression Data. Bioinformatics 18(1), 136–144 (2002)
Wu, C., Fu, Y., Murali, T.M., Kasif, S.: Gene Expression Module Discovery Using Gibbs Sampling. Genome Informatics 15(1), 239–248 (2004)
Madeira, S.C., Teixeira, M.C., Sá-Correia, L., Oliveira, A.L.: Identification of Regulatory Modules in Time Series Gene Expression Data Using a Linear Time Biclustering Algorithm. IEEE/ACM Transaction Computational Biology and Bioinformatics 7(1), 153–165 (2010)
Getz, G., Levine, E., Domany, E.: Coupled Two-Way Clustering Analysis of Gene Microarray Data. Proc. Natural Academy of Sciences Us, 12079–12084 (2000)
Madeira, S.C., Gonçalves, J.P., Oliveira, A.L.: Efficient Biclustering Algorithms for identifying transacriptional regulation relationships using time series gene expression data. INESC_ID Tec. Rep. 22 (2007)
Peeters, R.: The Maximum Edge Biclique Problem Is NP-Complete. Discrete Applied Math. 131(3), 651–654 (2003)
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2013 Springer International Publishing Switzerland
About this paper
Cite this paper
Phukhachee, T., Maneewongvatana, S. (2013). Identification of K-Tolerance Regulatory Modules in Time Series Gene Expression Data Using a Biclustering Algorithm. In: Yoshida, T., Kou, G., Skowron, A., Cao, J., Hacid, H., Zhong, N. (eds) Active Media Technology. AMT 2013. Lecture Notes in Computer Science, vol 8210. Springer, Cham. https://doi.org/10.1007/978-3-319-02750-0_15
Download citation
DOI: https://doi.org/10.1007/978-3-319-02750-0_15
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-02749-4
Online ISBN: 978-3-319-02750-0
eBook Packages: Computer ScienceComputer Science (R0)