Abstract
Cardiac ultrasound imaging requires a high frame rate in order to capture rapid motion. This can be achieved by multi-line acquisition (MLA), where several narrow-focused received lines are obtained from each wide-focused transmitted line. This shortens the acquisition time at the expense of introducing block artifacts. In this paper, we propose a data-driven learning-based approach to improve the MLA image quality. We train an end-to-end convolutional neural network on pairs of real ultrasound cardiac data, acquired through MLA and the corresponding single-line acquisition (SLA). The network achieves a significant improvement in image quality for both 5- and 7-line MLA resulting in a decorrelation measure similar to that of SLA while having the frame rate of MLA.
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Increasing the frame rate is a major challenge in 2D and 3D echocardiography. Investigating deformations at different stages of the cardiac cycle is crucial for cardiovascular imaging; hence high temporal resolution is highly desired in addition to the spatial resolution. There are several ways to increase the frame rate of ultrasound imaging; one of the most commonly used techniques, which is implemented in many ultrasound scanners, is multi-line acquisition (MLA) [1], often referred to as parallel receive beamforming (PRB) [2].
Single- vs. Multi-line Acquisition. In single-line acquisition (SLA), a narrow-focused pulse is transmitted by introducing transmit time delays through a linear phased array of acoustic transducer elements. Upon reception the obtained signal is dynamically focused along the receive (Rx) direction which is identical to the transmit (Tx) direction. The spatial region of interest is raster scanned line-by-line to obtain an ultrasound image.
The need to transmit a large number of pulses sequentially results in a low frame rate and renders SLA inadequate for cardiovascular imaging, where a high frame rate is mandatory, especially for quantitative analysis or during stress tests. For the same reason, SLA is neither useful for scanning large fields of view in real time 3D imaging applications.
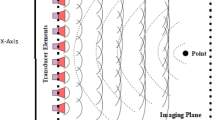
In an attempt to overcome the frame rate problem, the MLA method was proposed in [1, 3]. The main idea behind MLA is to transmit a weakly focused beam that provides a sufficiently wide coverage for a high number of received lines. On the receiver side, m lines is constructed from the data acquired from each transmit event, thereby increasing the frame rate by m (the latter number is usually referred to as the MLA factor). Signal formation in the SLA and MLA modalities is demonstrated in Fig. 1 where 5-MLA is depicted. For a 5-MLA, we construct 5 Rx lines per each Tx thus increasing the frame rate by the factor of 5.
MLA Artifacts. As the Tx and Rx are no longer aligned in the MLA mode, the two-way beam profile is shifted towards the original transmit direction, making the lateral sampling irregular [2]. This beam warping effect causes sharp lateral discontinuities that are manifested as block artifacts in the image domain.
The observed block artifacts in the ultrasound images (see, e.g., Fig. 1) tend to be more obvious when the number of transmit events decreases. The MLA artifact can be measured by assessing the correlation coefficient between each two adjacent Rx lines in the in-phase and quadrature (I/Q) demodulated beamformed data [4]. In SLA or compensated MLA, the averaged correlation values inside MLA groups and between MLA groups are almost the same. In the uncompensated cases, however, the correlation values are different.
Apart from beam warping, there are two other effects caused by the transmit-receive misalignment: skewing, where shape of the two-way beam profile becomes asymmetric, and gain variation, where the outermost lines inside the group have a lower gain than the innermost lines [4].
Related Work. Several methods have been proposed in literature to decrease MLA artifacts, including transmit sinc apodization [5] and dynamic steering [6], incoherent interpolation [7, 8] (applied after envelope detection), and its coherent (before envelope detection) counterparts [2, 9]. One of the more prominent methods, synthetic transmit beamforming (STB) [2], creates synthetic Tx lines by coherently interpolating information received from each two adjacent Tx events in intermediate directions. This technique creates highly correlated lines, attenuating block artifacts. A common practice for MLA imaging with focused beams is to create 2-4 Rx lines per each Tx event in cases without overlap, or 4-8 lines in the presence of overlaps from adjacent transmissions, in order to perform the correction [2, 4, 10]. Thus, creating eight lines with overlaps provides an effective frame rate increase by the factor of 4. In this paper, however, we used odd MLA factors \(m=5,7\) for the purpose of acquiring data from aligned directions for both SLA and MLA.
Recently, data-driven learning techniques based on convolutional neural networks (CNNs) have been extensively used for solving inverse problems in imaging and in medical imaging in particular, for example, in X-ray CT reconstruction and denoising [11]. Inspired by their success, we propose a data-driven approach to overcome MLA artifacts.
Contributions. We propose an end-to-end CNN-based approach for MLA artifact correction. Our fully convolutional network consists of interpolation layers followed by a trainable apodization layer, and is trained on in-vivo cardiac data to approximate an SLA quality image. We demonstrate the effectiveness of this network both visually and quantitatively using the decorrelation measure (\(D_c\)) and SSIM [12] quality criteria. To the best of our knowledge, this is the first study to report good artifact corrections in the case of 5-7-MLA. We show that the trained network generalizes well across patients, as well as to phantom data.
2 Methods
2.1 Improving MLA with CNNs
Aiming at providing a general and optimal solution for MLA interpolation achieving SLA quality, we propose to replace MLA artifact correction and apodization phases in the traditional MLA pipeline as shown in Fig. 2 with an end-to-end CNN depicted in Fig. 3. We draw similarities to [10] who showed that combining MLA interpolation with an optimal apodization method produces superior results compared to the traditional approaches. Our network comprises both the interpolation and the apodization stages that are trained jointly.
Interpolation Stage. The interpolation stage consists of our CNN containing 10 convolutional layers with symmetric skip connections [13, 14] from each layer in the downsampling track to its corresponding layer in the upsampling track as visualized in Fig. 3. Downsampling is performed using average pooling and strided convolutions are used for upsampling. The number of bifurcations is set to 5 for all the experiments. The interpolation stage takes as an input the time-delayed and phase-rotated element-wise I/Q data from the transducer.
Apodization Stage. Following the interpolation stage, we introduce a convolutional layer to perform apodization. This is performed using point-wise convolutions (\(1\times 1\)) for each element’s channel in the network and the results are then added to the learned weights of the convolution. The weights of the channel are initialized with a Hann window.
Optimization. We use the \(L_1\) norm training loss to measure the discrepancy between the image predicted by the network and the ground truth SLA images. The loss is minimized using the Adam optimizer [15] with a learning rate of \(10^{-4}\). We observed that adding the apodization stage accelerates the training process, and makes the network converge faster.
2.2 Data Acquisition and Training
We generated a dataset for training the network using cardiac data from six patients; each patient contributed 4-5 cine loops, containing 32 frames. The data was acquired using a GE experimental breadboard ultrasound system. The same transducer was used for both phantom and cardiac acquisition. Excitation sinusoidal pulses of 1.75 cycles, centered around 2.5 MHz, were transmitted using 28 central elements out of the total 64 element in the probe with a pitch of 0.3 mm, elevation size of 13 mm and elevation focus of 100 mm. The depth focus was set at 71 mm. In order to assess the desired aperture for MLA setup, Field II simulator [16] was used as in [10] using the transducer impulse response and tri-state transmission excitation sequence, requiring a minimal insonification of \(-3\) dB for all MLAs from a single Tx.
On the Rx side, the I/Q demodulated signals were dynamically focused using linear interpolation, with an f-number of 1. The FOV was covered with 140/140 Tx/ Rx lines in SLA mode, 28/140 Tx/Rx lines in the 5-MLA mode, and 20/140 Tx/Rx lines in the 7-MLA mode. For both phantom and cardiac cases, the data were acquired in the SLA mode; 5-MLA and 7-MLA data was obtained by appropriately decimating the Rx pre-beamformed data.
In total, we used 745 frames from five patients for training and validation, while keeping the cine loops from the sixth patient for testing. The data set comprised pairs of beamformed I/Q images with Hann window apodization, and the corresponding 5- and 7-MLA pre-apodization samples with the dimensions of \(652 \times 64 \times 140\) (depth \(\times \) elements \(\times \) Rx lines). The MLA data was acquired by decimation of the Tx lines of the SLA samples by the MLA factor (\(m=5,7\)).
We trained dedicated CNNs for the reconstruction of SLA images from 5- and 7-MLA. Each CNN was trained to a maximum of 200 epochs on mini batches of size 4.
3 Experimental Evaluation
3.1 Settings
In order to assess the performance of our trained networks, we used cine loops from one patient excluded from the training/validation set. From two cine loops, each containing 32 frames, we generated pairs of 5- and 7-MLA samples and their corresponding SLA images the same way as described in Sect. 2.2, resulting in 64 test samples. For quantitative evaluation of the performance of our method we measured the decorrelation (\(D_c\)) criterion that evaluates the artifact strength [4], and the SSIM [12] structural similarity criterion with respect to the SLA image. In addition, we tested the performance of our networks on four frames acquired from the GAMMEX Ultrasound 403GS LE Grey Scale Precision Phantom.
3.2 Results
Quantitative results for the cardiac test set are summarized in Table 1. We show a major improvement in decorrelation and SSIM for both 5- and 7-MLA. The corrected 7-MLA performance approaches that of 5-MLA, suggesting the feasibility of larger MLA factors. Figure 4 shows representative images from each imaging modality. We show that the correlation coefficients profile of the corrected 5- and 7-MLA approaches that of SLA.
Similarly, quantitative results for the phantom test set are summarized in Table 2, again showing a significant improvement in the image quality for both 5- and 7-MLA. Visual results with the corresponding correlation coefficients profiles are depicted in Fig. 1 in the Supplementary Material. These results suggest that the networks trained on real cardiac data generalize well to the phantom data without any further training or fine-tuning. For comparison, [4] reported a decorrelation value of \(-1.5\) for a phantom image acquired in a \(4-\)MLA mode with STB compensation, while we report closer to zero \(D_c\) values, 0.457 for 5-MLA and 0.956 for 7-MLA, which both use a greater decimation rate. The slight dissimilarities in the recovered data can be explained by the acquisition method being used: since the scanned object was undergoing a motion, there is a difference between all but a central line in each MLA group and the matching lines in SLA. We assume that training the network on images of static organs may further improve its performance. Independently, small areas with vertical stripes were observed in several images. In our opinion, the origin of the stripes is a coherent summation of the beamformed lines across the moving object. Since the frame rate of the employed acquisition sequence was slower than of genuine MLA acquisition, the magnitude of this artifact is probably exaggerated.
4 Conclusion
In this paper, we have shown that conventional ultrasound MLA correction can be substituted with an end-to-end CNN performing both optimal interpolation and apodization in order to approximate SLA image quality. In the future, we aim at extending this approach to even earlier stages in multi-line acquisition such as beamforming, assuming it will provide a greater improvement in image quality. Moreover, a similar method could probably be applied for other fast US acquisition modalities, such as multi-line transmission (MLT) [17].
References
Shattuck, D.P., Weinshenker, M.D., Smith, S.W., von Ramm, O.T.: Explososcan: a parallel processing technique for high speed ultrasound imaging with linear phased arrays. Acoust. Soc. Am. J. 75, 1273–1282 (1984)
Hergum, T., Bjastad, T., Kristoffersen, K., Torp, H.: Parallel beamforming using synthetic transmit beams. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 54(2), 271–280 (2007)
Ramm, O.T.V., Smith, S.W., Pavy, H.G.: High-speed ultrasound volumetric imaging system. II. parallel processing and image display. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 38(2), 109–115 (1991)
Bjastad, T., Aase, S.A., Torp, H.: The impact of aberration on high frame rate cardiac b-mode imaging. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 54(1), 32 (2007)
Augustine, L.J.: High resolution multiline ultrasonic beamformer. US Patent 4,644,795, 24 February 1987
Thiele, K.E., Brauch, A.: Method and apparatus for dynamically steering ultrasonic phased arrays. US Patent 5,322,068, 21 June 1994
Holley, G.L., Guracar, I.M.: Ultrasound multi-beam distortion correction system and method. US Patent 5,779,640, 14 July 1998
Liu, D.D., Lazenby, J.C., Banjanin, Z., McDermott, B.A.: System and method for reduction of parallel beamforming artifacts. US Patent 6,447,452, 10 September 2002
Wright, J.N., Maslak, S.H., Finger, D.J., Gee, A.: Method and apparatus for coherent image formation. US Patent 5,623,928, 29 April 1997
Rabinovich, A., Friedman, Z., Feuer, A.: Multi-line acquisition with minimum variance beamforming in medical ultrasound imaging. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 60(12), 2521–2531 (2013)
McCann, M.T., Jin, K.H., Unser, M.: Convolutional neural networks for inverse problems in imaging: a review. IEEE Signal Process. Mag. 34(6), 85–95 (2017)
Wang, Z., Bovik, A.C., Sheikh, H.R., Simoncelli, E.P.: Image quality assessment: from error visibility to structural similarity. IEEE Trans. Image Process. 13(4), 600–612 (2004)
Mao, X., Shen, C., Yang, Y.B.: Image restoration using very deep convolutional encoder-decoder networks with symmetric skip connections. In: Advances in Neural Information Processing Systems, pp. 2802–2810 (2016)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Kingma, D.P., Ba, J.: Adam: a method for stochastic optimization. In: Proceedings of the 3rd International Conference on Learning Representations (ICLR) (2015)
Jensen, J.A.: Field: a program for simulating ultrasound systems. In: 10th Nordicbaltic Conference on Biomedical Imaging, vol. 4, Supplement 1, Part 1, pp. 351–353. Citeseer (1996)
Mallart, R., Fink, M.: Improved imaging rate through simultaneous transmission of several ultrasound beams. In: New Developments in Ultrasonic Transducers and Transducer Systems, vol. 1733, pp. 120–131. International Society for Optics and Photonics (1992)
Acknowledgements
The research was partially supported by ERC StG RAPID.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
1 Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this paper
Cite this paper
Senouf, O. et al. (2018). High Frame-Rate Cardiac Ultrasound Imaging with Deep Learning. In: Frangi, A., Schnabel, J., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds) Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. MICCAI 2018. Lecture Notes in Computer Science(), vol 11070. Springer, Cham. https://doi.org/10.1007/978-3-030-00928-1_15
Download citation
DOI: https://doi.org/10.1007/978-3-030-00928-1_15
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-00927-4
Online ISBN: 978-3-030-00928-1
eBook Packages: Computer ScienceComputer Science (R0)