Abstract
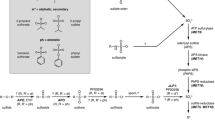
Pseudomonas species inhabit a wide variety of habitats, ranging from the human body to soils, the rhizosphere, and the phyllosphere. Like all microorganisms, they require sulfur for growth, and this is normally provided by assimilation of inorganic sulfate. However, many pseudomonads inhabit environments where sulfate may not be freely available. In aerobic soils, for instance, inorganic sulfate makes up less than 5% of the total sulfur, and most of the residual sulfur is present as peptides/amino acids, sulfate esters, and sulfonates. Much of this sulfonate content is derived from plant sulfolipid in the thylakoid membranes, which may also provide sulfur for leaf-dwelling pseudomonads. In the human body, other sulfur sources are present, such as the neurotransmitter taurine (2-aminoethane-sulfonate), sulfated mucins in gut and lung environments, and glycosaminoglycan in connective tissue. The importance of sulfur metabolism for pseudomonads is underlined by the fact that in Pseudomonas syringae methionine prototrophy is required for virulence8, while in Pseudomonas putida loss of the ability to desulfurize sulfate esters leads to reduced survival in the soil54.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
Alaminos, M. and Ramos, J.L., 2001, The methionine biosynthetic pathway from homoserine in Pseudomonas putida involves the metW, metX, metZ, metH and metE gene products. Arch. Microbiol., 176:151–154.
Andersen, G.L., Beattie, G.A., and Lindow, S.E., 1998, Molecular characterization and sequence of a methionine biosynthetic locus from Pseudomonas syringae. J. Bacteriol., 180:4497–4507.
Autry, A.R. and Fitzgerald, J.W., 1990, Sulfonate S—A major form of forest soil organic sulfur. Biol. Fertil. Soils, 10:50–56.
Baek, M.C., Kim, S.K., Kim, D.H., Kim, B.K., and Choi, E.C., 1996, Cloning and sequencing of the Klebsiella K-36 astA gene, encoding an arylsulfate sulfotransferase. Micmbiol. Immunol., 40:531–537.
Baker, S.C., Kelly, D.P., and Murrell, J.C., 1991, Microbial degradation of methanesulfonic acid—a missing link in the biogeochemical sulfur cycle. Nature, 350:627–628.
Bateman, T.J., Dodgson, K.S., and White, G.F., 1986, Primary alkylsulfatase activities of the detergent-degrading bacterium Pseudomonas C12B: Purification and properties of the P1 enzyme. Biochem. J., 236:401–408.
Beattie, G.A. and Lindow, S.E., 1994, Comparison of the behavior of epiphytic fitness mutants of Pseudomonas syringae under controlled and field conditions. Appl. Environ. Micmbiol., 60:3799–3808.
Beattie, G.A. and Lindow, S.E., 1994, Survival, growth, and localization of epiphytic fitness mutants of Pseudomonas syringae on leaves. Appl. Environ. Micmbiol., 60:3790–3798.
Beil, S., Kehrli, H., James, P., Staudenmann, W., Cook, A.M., Leisinger, T., and Kertesz, M.A., 1995, Purification and characterization of the arylsulfatase synthesized by Pseudomonas aeruginosa PAO during growth in sulfate-free medium and cloning of the arylsulfatase gene (atsA). Eur. J. Biochem., 229:385–394
Bick, J.A., Dennis, J.J., Zylstra, G.J., Nowack, J., and Leustek, T., 2000, Identification of a new class of 5′-adenylylsulfate (APS) reductases from sulfate-assimilating bacteria. J. Bacteriol., 182:135–142.
Boltes, I., Czapinska, H., Kahnert, A., von Bülow, R., Dierks, T., Schmidt, B., von Figura, K., Kertesz, M.A., and Usón, I., 2001,1.3 Å crystal structure of arylsulfatase from Pseudomonas aeruginosa establishes the catalytic mechanism for sulfate ester cleavage in the sulfatase family. Structure, 9:483–491
Bond, C.S., Clements, P.R., Ashby, S.J., Collyer, CA., Harrop, S.J., Hopwood, J.J., and Guss, J.M., 1997, Structure of a human lysosomal sulfatase. Structure, 5:277–289.
Brilon, C., Beckmann, W., Hellwig, M., and Knackmuss, H.J., 1981, Enrichment and isolation of naphthalenesulfonic acid-utilizing pseudomonads. Appl. Environ. Microbiol., 42:39–43.
Bykowski, T., van der Ploeg, J.R., Iwanicka-Nowicka, R., and Hryniewicz, M.M., 2002, The switch from inorganic to organic sulphur assimilation in Escherichia coli: Adenosine 5′-phosphosulphate (APS) as a signalling molecule for sulphate excess. Mol. Microbiol., 43:1347–1358.
Campos-García, J., Esteve, A., Vázquez-Duhalt, R., Ramos, J.L., and Soberón-Chàvez, G., 1999, The branched-chain dodecylbenzene sulfonate degradation pathway of Pseudomonas aeruginosa W51D involves a novel route for degradation of the surfactant lateral alkyl chain. Appl. Environ. Microbiol., 65:3730–3734.
Chai, C.L.L. and Lowe, G., 1992, The mechanism and stereochemical course of sulfuryl transfer catalyzed by the aryl sulfotransferase from Eubacterium A-44. Bioorg. Chem., 20:181–188.
Chance, D.L. and Mawhinney, T.P., 2000, Carbohydrate sulfation effects on growth of Pseudomonas aeruginosa. Microbiology, 146:1717–1725.
Clinch, K., Evans, G.B., Furneaux, R.H., Rendle, P.M., Rhodes, PL., Robertson, A.M., Rosendale, D.I., Tyler, P.C., and Wright, D.P., 2002, Synthesis and utility of sulfated chromogenic carbohydrate model substrates for measuring activities of mucin-desulfating enzymes. Carbohydr. Res., 337:1095–1111.
Cloves, J.M., Dodgson, K.S., White, G.F., and Fitzgerald, J., 1980, Purification and properties of the P-2 primary alkylsulfohydrolase of the detergent-degrading bacterium Pseudomonas C-12B. Biochem. J., 185:23–32.
Cook, A.M., Laue, H., and Junker, F., 1999, Microbial desulfonation. FEMS Microbiol. Rev., 22:399–419.
Davison, J., Brunei, F., Phanopoulos, A., Prozzi, D., and Terpstra, P., 1992, Cloning and sequencing of Pseudomonas genes determining sodium dodecyl sulfate biodegradation. Gene, 114:19–24.
Delic-Attree, L, Toussaint, B., Garin, J., and Vignais, P.M., 1997, Cloning, sequence and mutagenesis of the structural gene of Pseudomonas aeruginosa CysB, which can activate algD transcription. Mol. Microbiol., 24:1275–1284.
Delisle, G. and Milazzo, F.H., 1970, The isolation of arylsulphatase isoenzymes from Pseudomonas aeruginosa. Biochim. Biophys. Acta, 212:505–508.
Delisle, G.J. and Milazzo, F.H., 1972, Characterization of arylsulfatase isoenzymes from Pseudomonas aeruginosa. Can. J. Microbiol., 18:561–568.
Denger, K. and Cook, A.M., 2001, Ethanedisulfonate is degraded via sulfoacetaldehyde in Ralstonia sp. strain EDS1. Arch. Microbiol., 176:89–95.
Dierks, T., Miech, C., Hummerjohann, J., Schmidt, B., Kertesz, M.A., and von Figura, K., 1998, Posttranslational formation of formylglycine in prokaryotic sulfatases by modification of either cysteine or serine. J. Biol. Chem., 273:25560–25564.
Dodgson, K.S., White, G.F., and Fitzgerald, J.W., 1982, Sulfatases of microbial origin. CRC Press, Boca Raton.
Eaton, R.W. and Nitterauer, J.D., 1994, Biotransformation of benzothiophene by isopropyl-benzene-degrading bacteria. J. Bacteriol., 176:3992–4002.
Eichhorn, E., van der Ploeg, J.R., Kertesz, M.A., and Leisinger, T., 1997, Characterization of α-ketoglutarate dependent taurine dioxygenase from Escherichia coli. J. Biol. Chem., 272:23031–23036.
Eichhorn, E., van der Ploeg, J.R., and Leisinger, T., 1999, Characterization of a two-component alkanesulfonate monooxygenase from Escherichia coli. J. Biol. Chem., 274:26639–26646.
Eichhorn, E., van der Ploeg, J.R., and Leisinger, T., 2000, Deletion analysis of the Escherichia coli taurine and alkanesulfonate transport systems. J. Bacteriol., 182:2687–2695.
Elkins, J.M., Ryle, M.J., Clifton, I.J., Hotopp, J.C.D., Lloyd, J.S., Burzlaff, N.I., Baldwin, IE., Hausinger, R.P., and Roach, PL., 2002, X-ray crystal structure of Escherichia coli taurine/alpha-ketoglutarate dioxygenase complexed to ferrous iron and substrates. Biochemistry, 41:5185–5192.
Endoh, T., Habe, H., Yoshida, T., Nojiri, H., and Omori, T., 2003, A CysB-regulated and sigma-54-dependent regulator, SfnR, is essential for dimethyl sulfone metabolism of Pseudomonas putida strain DS1. Microbiology, 149:991–1000.
Endoh, T., Kasuga, K., Horinouchi, M., Yoshida, T., Habe, H., Nojiri, H., and Omori, T., 2003, Characterization and identification of genes essential for dimethyl sulfide utilization in Pseudomonas putida strain DS1 Appl. Microbiol. Biotechnol., 62:83–91.
Fitzgerald, J.W., Dodgson, K.S., and Payne, W.J., 1974, Induction of primary alkylsulphatases and metabolism of sodium hexan-1-yl sulphate by Pseudomonas C12B. Biochem. J., 138:63–69.
Fitzgerald, J.W. and Payne, W.J., 1972, The regulation of arylsulfatase activity in Pseudomonas C12B. Microbios., 6:147–156.
Fitzgerald, J.W. and Scott, C.L., 1974, Utilization of choline-0-sulphate as a sulphur source for growth by a Pseudomonas isolate. Microbios, 10:121–131.
Fitzgerald, J.W., Stewart, G.J., and Kight Olliff, L., 1980, Regulation of primary alkylsulfatase induction in Pseudomonas C12B: Concentration-dependent stimulation-inhibition by exogenous UTP and sodium acetate and inhibition by 1-hexanol. Can. J. Microbiol., 26:1348–1355.
Foglino, M., Borne, F., Bally, M., Ball, G., and Patte, J.C., 1995, A direct sulfhydrylation pathway is used for methionine biosynthesis in Pseudomonas aeruginosa. Microbiology, 141:431–439.
Gallardo, M.E., Ferrandez, A., De Lorenzo, V., Garcia, J.L., and Diaz, E., 1997, Designing recombinant Pseudomonas strains to enhance biodesulfurization. J. Bacteriol., 179:7156–7160.
Gray, K.A., Pogrebinsky, O.S., Mrachko, G.T., Xi, L., Monticello, DJ., and Squires, C.H., 1996, Molecular mechanisms of biocatalytic desulfurization of fossil fuels. Nature Biotechnol., 14:1705–1709.
Günther, E., Petruschka, L., and Herrmann, H., 1979, Reverse transsulfuration pathway in Pseudomonas aeruginosa. Z. Allg. Mikrobiol., 19:439–442.
Harada, T., 1964, The formation of sulphatases in Pseudomonas aeruginosa. Biochim. Biophys. Acta, 81:193–196.
Haug, W., Schmidt, A., Nortemann, B., Hempel, D.C., Stolz, A., and Knackmuss, H.J., 1991, Mineralization of the sulfonated azo dye Mordant Yellow-3 by a 6-aminonaphthalene-2-sulfonate-degrading bacterial consortium. Appl. Environ. Microbiol., 57: 3144–3149.
Hsu, Y.C., 1965, Detergent-splitting enzyme from Pseudomonas. Nature, 207:385–388.
Hummerjohann, J., Kuttel, E., Quadroni, M., Ragaller, J., Leisinger, T., and Kertesz, M.A., 1998, Regulation of the sulfate starvation response in Pseudomonas aeruginosa: Role of cysteine biosynthetic intermediates. Microbiology, 144:1375–1386.
Hummerjohann, I, Laudenbach, S., Rétey, J., Leisinger, T., and Kertesz, M.A., 2000, The sulfur-regulated arylsulfatase gene cluster of Pseudomonas aeruginosa, a new member of the cys regulon. J. Bacteriol., 182:2055–2058.
Imperato, T.J., Wong, CG., Chen, L.J., and Bolt, R.J., 1977, Hydrolysis of lithocholate sulfate by Pseudomonas aeruginosa. J. Bacteriol., 130:545–547.
Inoue, H., Inagaki, K., Eriguchi, S.I., Tamura, T., Esaki, N., Soda, K., and Tanaka, H., 1997, Molecular characterization of the mde operon involved in L-methionine catabolism of Pseudomonas putida. J. Bacteriol., 179:3956–3962.
Inoue, H., Inagaki, K., Sugimoto, M., Esaki, N., Soda, K., and Tanaka, H., 1995, Structural analysis of the L-methionine gamma-lyase gene from Pseudomonas putida. J. Biochem., 117:1120–1125.
Jansen, HJ., Hart, CA., Rhodes, J.M., Saunders, J.R., and Smalley, J.W., 1999, A novel mucin-sulphatase activity found in Burkholderia cepacia and Pseudomonas aeruginosa. J. Med. Microbiol., 48:551–557.
Junker, F., Leisinger, T., and Cook, A.M., 1994, 3-Sulphocatechol 2,3-dioxygenase and other dioxygenases (EC 1.13.11.2 and EC 1.14.12.-) in the degradative pathways of 2-amino-benzenesulphonic, benzenesulphonic and 4-toluenesulphonic acids in Alcaligenes sp. strain O-1. Microbiology, 140:1713–1722.
Kahnert, A. and Kertesz, M.A., 2000, Characterization of a sulfur-regulated oxygenative alkylsulfatase from Pseudomonas putida S-313. J. Biol. Chem., 275:31661–31667.
Kahnert, A., Mirleau, P., Wait, R., and Kertesz, M.A., 2002, The LysR-type regulator SftR is involved in soil survival and sulfate ester metabolism in Pseudomonas putida. Environ. Microbiol., 4:225–237.
Kahnert, A., Vermeij, P., Wietek, C., James, P., Leisinger, T., and Kertesz, M.A., 2000, The ssu locus plays a key role in organosulfur metabolism in Pseudomonas putida S-313. J. Bacteriol., 182:2869–2878.
Kang, J.W., Kwon, A.R., Kim, D.H., and Choi, E.C., 2001, Cloning and sequencing of the astA gene encoding arylsulfate sulfotransferase from Salmonella typhimurium. Biol. Pharm. Bull., 24:570–574.
Kertesz, M.A., 2001, Bacterial transporters for sulfate and organosulfur compounds. Res. Microbiol., 152:279–290.
Kertesz, M.A., 1996, Desulfonation of aliphatic sulfonates by Pseudomonas aeruginosa PAO. FEMS Microbiol. Lett., 137:221–225.
Kertesz, M.A., 1999, Riding the sulfur cycle—metabolism of sulfonates and sulfate esters in Gram-negative bacteria. FEMS Microbiol. Rev., 24:135–175.
Kertesz, M.A., Kölbener, P., Stockinger, H., Beil, S., and Cook, A.M., 1994, Desulfonation of linear alkylbenzenesulfonate surfactants and related compounds by bacteria. Appl. Environ. Microbiol., 60:2296–2303.
Kertesz, M.A., Leisinger, T., and Cook, A.M., 1993, Proteins induced by sulfate limitation in Escherichia coli, Pseudomonas putida, or Staphylococcus aureus. J. Bacteriol., 175:1187–1190.
Kertesz, M.A., Schmidt-Larbig, K., and Wüest, T., 1999, A novel reduced flavin mononucleotide-dependent methanesulfonate sulfonatase encoded by the sulfur-regulated msu operon of Pseudomonas aeruginosa. J. Bacteriol., 181:1464–1473.
Key, B.D., Howell, R.D., and Griddle, C.S., 1998, Defluorination of organofluorine sulfur compounds by Pseudomonas sp. strain D2. Environ. Sci. Technol., 32:2283–2287.
King, J.E., Jaouhari, R., and Quinn, J.P., 1997, The role of sulfoacetaldehyde sulfo-lyase in the mineralization of isethionate by an environmental Acinetobacter isolate. Microbiology, 7:2339–2343.
King, J.E. and Quinn, J.P., 1997, Metabolism of sulfoacetate by environmental Aureobacterium sp. and Comamonas acidovorans isolates. Microbiology, 12:3907–3912.
Kopriva, S., Buchert, T., Fritz, G., Suter, M., Benda, R.D., Schunemann, V., Koprivova, A., Schurmann, P., Trautwein, A.X., Kroneck, P.M.H., and Brunold, C., 2002, The presence of an iron-sulfur cluster in adenosine 5′-phosphosulfate reductase separates organisms utilizing adenosine 5′-phosphosulfate and phosphoadenosine 5′-phosphosulfate for sulfate assimilation. J. Biol. Chem., 277:21786–21791.
Kredich, N.M., 1996, Biosynthesis of cysteine. In F.C. Neidhardt, R. Curtiss, J.L. Ingraham, E.C.C. Lin, K.B. Low, B. Magasanik, W.S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (eds), Escherichia coli and Salmonella typhimurium. Cellular and Molecular Biology, 2nd edn, pp. 514–527. ASM Press, Washington.
Kropp, K.G. and Fedorak, P.M., 1998, A review of the occurrence, toxicity, and biodegradation of condensed thiophenes found in petroleum. Can. J. Microbiol., 44:605–622.
Kwon, A.R., Yun, H.J., and Choi, E.C., 2001, Kinetic mechanism and identification of the active site tyrosine residue in Enterobacter amnigenus arylsulfate sulfotransferase. Biochem. Biophys. Res. Commun., 285:526–529.
Lee, N.A. and Clark, D.P., 1993, A natural isolate of Pseudomonas maltophila which degrades aromatic sulfonic acids. FEMS Microbiol. Lett., 107:151–155.
Lisa, T.A., Casale, C.H., and Domenech, C.E., 1994, Cholinesterase, acid phosphatase, and phospholipase C of Pseudomonas aeruginosa under hyperosmotic conditions in a high phosphate medium. Curr. Microbiol., 28:71–76.
Locher, H.H., Leisinger, T., and Cook, A.M., 1991, 4-Sulphobenzoate 3,4-dioxygenase. Purification and properties of a desulphonative two-component enzyme system from Comamonas testosteroni T-2. Biochem. J., 274:833–842.
Lucas, J.J., Burchiel, S.W., and Segel, I.H., 1972, Choline sulfatase of Pseudomonas aeruginosa. Arch. Biochem. Biophys., 153:664–672.
Lukatela, G., Krauss, N., Theis, K., Selmer, T., Gieselmann, V., von Figura, K., and Saenger, W., 1998, Crystal structure of human arylsulfatase A: The aldehyde function and the metal ion at the active site suggest a novel mechanism for sulfate ester hydrolysis. Biochemistry, 37:3654–3664.
Martelli, H.L. and Souza, S.M., 1970, Biochemistry of sulfonic compounds III. Formation of a two-carbon compound during the oxidation of sulfoacetate by a Pseudomonas strain. Biochim. Biophys. Acta, 208:110–115.
Matcham, G.W.J., Bartholomew, B., Dodgson, K.S., Fitzgerald, J.W., and Payne, W.J., 1977, Stereospecificity and complexity of microbial sulphohydrolases involved in the biodegradation of secondary alkylsulphate detergents. FEMS Microbiol. Lett., 1:197–200.
McFarland, B.L., 1999, Biodesulfurization. Curr. Opin. Microbiol., 2:257–264.
Miech, C., Dierks, T., Selmer, T., von Figura, K., and Schmidt, B., 1998, Arylsulfatase from Klebsiella pneumoniae carries a formylglycine generated from a serine. J. Biol. Chem., 273:4835–4837.
Monticello, D.J., 2000, Biodesulfurization and the upgrading of petroleum distillates. Curr. Opin. Biotechnol., 11:540–546.
Murooka, Y., Ishibashi, K., Yasumoto, M., Sasaki, M., Sugino, H., Azakami, H., and Yamashita, M., 1990, A sulfur-and tyramine-regulated Klebsiella aerogenes operon containing the arylsulfatase (atsA) gene and the atsB gene. J. Bacteriol., 172:2131–2140.
Nelson, J.W, Tredgett, M.W., Sheehan, J.K., Thornton, DJ., Notman, D., and Govan, J.R.W., 1990, Mucinophilic and chemotactic properties of Pseudomonas aeruginosa in relation to pulmonary colonization in cystic fibrosis. Infect. Immun., 58:1489–1495.
Nelson, K.E., Weinel, C., Paulsen, I.T., Dodson, R.J., Hilbert, H., dos Santos, V., Fouts, D.E., Gill, S.R., Pop, M., Holmes, M., Brinkac, L., Beanan, M., DeBoy, R.T., Daugherty, S., Kolonay, J., Madupu, R., Nelson, W., White, O., Peterson, J., Khouri, H., Hance, I., Lee, P.C., Holtzapple, E., Scanlan, D., Tran, K., Moazzez, A., Utterback, T., Rizzo, M., Lee, K., Kosack, D., Moestl, D., Wedler, H., Lauber, J., Stjepandic, D., Hoheisel, J., Straetz, M., Heim, S., Kiewitz, C., Eisen, J., Timmis, K.N., Dusterhoft, A., Tummler, B., and Fraser, CM., 2002, Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ. Microbiol., 4:799–808.
Ochsner, U.A. and Vasil, M.L., 1996, Gene repression by the ferric uptake regulator in Pseudomonas aeruginosa: Cycle selection of iron-regulated genes. Proc. Natl. Acad. Sci. USA, 93:4409–4414.
Ohe, T., Ohmoto, T., Kobayashi, Y., Sato, A., and Watanabe, Y., 1990, Metabolism of naph-thalenesulfonic acids by Pseudomonas sp. TA-2. Agric. Biol. Chem., 54:669–675.
Ohe, T. and Watanabe, Y., 1986, Degradation of 2-naphthylamine-l-sulfonic acid by Pseudomonas strain TA-1. Agric. Biol. Chem., 50:1419–1426
Ohe, T. and Watanabe, Y., 1988, Microbial degradation of 1,6-naphthalenedisulfonic and 2,6-naphthalenedisulfonic acid by Pseudomonas sp. DS-1. Agric. Biol. Chem., 52:2409–2414.
Osteras, M., Boncompagni, E., Vincent, N., Poggi, M.C., and Le Rudulier, D., 1998, Presence of a gene encoding choline sulfatase in Sinorhizobium meliloti bet operon: Choline-O-sulfate is metabolized into glycine betaine. Proc. Natl. Acad. Sci. USA, 95:11394–11399.
Pao, S.S., Paulsen, LT., and Saier, M.H., Jr, 1998, Major facilitator superfamily. Microbiol. Mol. Biol. Rev., 62:1–34.
Payne, W.J. and Faisal, YE., 1963, Bacterial utilization of dodecylsulfate and dodecyl benzenesulfonate. Appl. Microbiol., 11:339–344.
Proksova, M., Augustin, J., and Vrbanova, A., 1997, Enrichment, isolation and characterization of dialkyl sulfosuccinate degrading bacteria Comamonas terrigena N3H and Comamonas terrigena N1C. Folia Microbiol., 42:635–639.
Quadroni, M., James, P., Dainese-Hatt, P., and Kertesz, M.A., 1999, Proteome mapping, mass spectrometric sequencing and reverse transcriptase-PCR for characterisation of the sulfate starvation-induced response in Pseudomonas aeruginosa PAO1. Eur. J. Biochem., 266:986–996.
Quadroni, M., Staudenmann, W., Kertesz, M., and James, P., 1996, Analysis of global responses by protein and peptide fingerprinting of proteins isolated by two-dimensional gel electrophoresis: Application to the sulfate-starvation response of Escherichia coli Eur. J. Biochem., 239:773–781.
Quick, A., Russell, N.J., Hales, S.G., and White, G.F., 1994, Biodegradation of sulphosuccinate: Direct desulphonation of a secondary sulphonate. Microbiology, 140:2991–2998.
Rammler, D.H., Grado, C., and Fowler, L.R., 1964, Sulfur metabolism of Aerobacter aerogenes 1. A repressible sulfatase. Biochemistry, 3:224–230.
Ramphal, R., Carnoy, C., Fievre, S., Michalski, J.C., Houdret, N., Lamblin, G., Strecker, G., and Roussel, P., 1991, Pseudomonas aeruginosa recognizes carbohydrate chains containing type 1 (Gal-Beta-l-3GlcNac) or Type-2 (Gal-Beta-l-4GlcNac) disaccharide units. Infect. Immun., 59:700–704.
Reichenbecher, W., Kelly, D.P., and Murreil, J.C., 1999, Desulfonation of propanesulfonic acid by Comamonas acidovorans strain P53: Evidence for an alkanesulfonate sulfonatase and an atypical sulfite dehydrogenase. Arch. Microbiol., 172:387–392.
Reichenbecher, W. and Murreil, J.C., 1999, Linear alkanesulfonates as carbon and energy sources for gram-positive and gram-negative bacteria. Arch. Microbiol., 171:430–438.
Rocha, E.P. C., Sekowska, A., and Danchin, A., 2000, Sulphur islands in the Escherichia coli genome: Markers of the cell’s architecture? FEBS Lett., 476:8–11.
Ruff, J., Hitzler, T., Rein, U., Ritter, A., and Cook, A.M., 1999, Bioavailability of water-polluting sulfonoaromatic compounds. Appl Microbiol. Biotechnol., 52:446–450.
Satishchandran, C., Hickman, Y.N., and Markham, G.D., 1992, Characterization of the phosphorylated enzyme intermediate formed in the adenosine 5′-phosphosulfate kinase reaction. Biochemistry, 31:11684–11688.
Schleheck, D., Dong, W.B., Denger, K., Heinzle, E., and Cook, A.M., 2000, An alpha-proteobacterium converts linear alkylbenzenesulfonate surfactants into sulfophenylcarboxylates and linear alkyldiphenyletherdisulfonate surfactants into sulfodiphenylethercar-boxylates. Appl. Environ. Microbiol., 66:1911–1916.
Schulz, S., Dong, W.B., Groth, U., and Cook, A.M., 2000, Enantiomeric degradation of 2-(4-sulfophenyl) butyrate via 4-sulfocatechol in Delftia acidovorans SPB1. Appl. Environ. Microbiol., 66:1905–1910.
Seitz, A.P., Leadbetter, E.R., and Godchaux, W., 1993, Utilization of sulfonates as sole sulfur source by soil bacteria including Comamonas acidovorans. Arch. Microbiol., 159:440–444.
Shan, X.Q., Chen, B., Zhang, T.H., Li, F.L., Wen, B., and Qian, J., 1997, Relationship between sulfur speciation in soils and plant availability. Sci. Total Environ., 199:237–246.
Shaw, D.J., Dodgson, K.S., and White, G.F., 1980, Substrate specificity and other properties of the inducible S3 secondary alkylsulphohydrolase purified from the detergent-degrading bacterium Pseudomonas C12B. Biochem. J., 187:181–196.
Shimamoto, G. and Berk, R.S., 1979, Catabolism of taurine in Pseudomonas aeruginosa. Biochim. Biophys. Acta, 569:287–292.
Shimamoto, G. and Berk, R.S., 1980, Taurine catabolism II. Biochemical and genetic evidence for sulfoacetaldehyde sulfolyase involvement. Biochim. Biophys. Acta, 632:121–130.
Smith, L.T., Pocard, J.A., Bernard, T., and Le Rudulier, D., 1988, Osmotic control of glycine betaine biosynthesis and degradation in Rhizobium meliloti. J. Bacteriol., 170:3142–3149.
Stolz, A., 2001, Basic and applied aspects in the microbial degradation of azo dyes. Appl. Microbiol. Biotechnol., 56:69–80.
Stolz, A., 1999, Degradation of substituted naphthalenesulfonic acids by Sphingomonas xenophaga BN6. J. Ind. Microbiol. Biotechnol., 23:391–399.
Stover, C.K., Pham, X.Q., Erwin, A.L., Mizoguchi, S.D., Warrener, P., Hickey, M.J., Brinkman, F.S.L., Hufhagle, W.O., Kowalik, DJ., Lagrou, M., Garber, R.L., Goltry, L., Tolentino, E., Westbrock-Wadman, S., Yuan, Y., Brody, L.L., Coulter, S.N., Folger, K.R., Kas, A., Larbig, K., Lim, R., Smith, K., Spencer, D, Wong, G.K.S., Wu, Z., Paulsen, LT., Reizer, J., Saier, M.H., Hancock, R.E.W, Lory, S., and Olson, M.V., 2000, Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature, 406:959–964.
Strous, G.J. and Dekker, J., 1992, Mucin-type glycoproteins. Crit. Rev. Biochem. Mol. Biol., 27:57–92.
Szameit, C., Miech, C., Balleininger, M., Schmidt, B., von Figura, K., and Dierks, T., 1999, The iron sulfur protein AtsB is required for posttranslational formation of formylglycine in the Klebsiella sulfatase. J. Biol. Chem., 274:15375–15381.
Tazuke, Y., Matsuda, K., Adachi, K., and Tsukada, Y., 1998, Purification and properties of a novel sulfatase from Pseudomonas testosteroni that hydrolyzed 3 beta-hydroxy-5-cholenoic acid 3-sulfate. Biosci. Biotechnol. Biochem., 62:1739–1744.
Tazuke, Y., Matsuda, K., Adachi, K., and Tsukada, Y., 1994, Purification and properties of bile acid sulfate sulfatase from Pseudomonas testosteroni. Biosci. Biotechnol. Biochem., 58:889–894.
Tazuke, Y., Matsuda, K., Okada, S., and Tsukada, Y., 1992, A novel sulfatase from Pseudomonas testosteroni hydrolyzing lithocholic acid sulfate. Biosci. Biotechnol. Biochem., 56:1584–1588.
Thomas, O.R.T., Matts, P.J., and White, G.F., 1988, Localization of electron microscopy of alkylsulfatases in bacterial cells. J. Gen. Microbiol., 134:1229–1236.
Thysse, G.J.E. and Wanders, T.H., 1972, Degradation of n-alkane-1-sulfonates by Pseudomonas. Antonie van Leeuwenhoek, 38:53–63.
Thysse, G.J.E. and Wanders, T.H., 1974, Initial steps in the degradation of n-alkane-1-sulphonates by Pseudomonas. Antonie van Leeuwenhoek, 40:25–37.
Tralau, T., Cook, A.M., and Ruff, J., 2001, Map of the IncPl beta plasmid pTSA encoding the widespread genes (tsa) for P-toluenesulfonate degradation in Comamonas testosteroniT-2. Appl. Environ. Microbiol., 67:1508–1516.
Tralau, T., Mampel, J., Cook, A.M., and Ruff, J., 2003, Characterization of TsaR, an oxygensensitive LysR-type regulator for the degradation of p-toluenesulfonate in Comamonas testosteroni T-2. Appl Environ Microbiol., 69:2298–2305.
Tralau, T., Wietek, C., and Kertesz, M.A., 2003, Desulfonation of aliphatic and aromatic sulfonates in Pseudomonas putida S-313 by a sulfate starvation-induced monooxygenase system, unpublished results.
Tsai, H.H., Hart, CA., and Rhodes, J.M., 1991, Production of mucin degrading sulphatases and glycosidases by Bacteroides thetaiotaomicron. Lett. Appl Microbiol., 13:97–101.
Vairavamurthy, M.A., Maletic, D., Wang, S.K., Manowitz, B., Eglinton, T., and Lyons, T., 1997, Characterization of sulfur-containing functional groups in sedimentary humic substances by X-ray absorption near-edge structure spectroscopy. Energy Fuels, 11:546–553.
Vairavamurthy, M.A., Zhou, W., Eglinton, T., and Manowitz, B., 1994, Sulfonates: A new class of organic sulfur compounds in marine sediments. Geochim. Cosmochim. Acta, 58:4681–4687.
van der Ploeg, J.R., Cummings, N.J., Leisinger, T., and Connerton, LE, 1998, Bacillus subtilis genes for the utilization of sulfur from aliphatic sulfonates. Microbiology, 144:2555–2561
van der Ploeg, J.R., Eichhorn, E., and Leisinger, T., 2001, Sulfonate-sulfur metabolism and its regulation in Escherichia coli Arch. Microbiol., 176:1–8.
van der Ploeg, J.R., Iwanicka-Nowicka, R., Bykowski, T., Hryniewicz, M., and Leisinger, T., 1999, The Cbl-regulated ssuEADCB gene cluster is required for aliphatic sulfonate-sulfur utilization in Escherichia coli J. Biol. Chem., 174:29358–29365
van der Ploeg, J.R., Iwanicka-Nowicka, R., Kertesz, M.A., Leisinger, T., and Hryniewicz, M.M., 1997, Involvement of CysB and Cbl regulatory proteins in expression of the tauABCD operon and other sulfate starvation-inducible genes in Escherichia coli J. Bacteriol., 179:7671–7678.
Vermeij, P. and Kertesz, M.A., 1999, Pathways of assimilative sulfur metabolism in Pseudomonas putida. J. Bacteriol., 181:5833–5837.
Vermeij, P., Wietek, C., Kahnert, A., Wüest, T., and Kertesz, M.A., 1999, Genetic organization of sulfur-controlled aryl desulfonation in Pseudomonas putida S-313. Mol. Microbiol., 32:913–926.
von Figura, K., Schmidt, B., Selmer, T., and Dierks, T., 1998, A novel protein modification generating an aldehyde group in sulfatases: Its role in catalysis and disease. Bioessays, 20:505–510.
Watwood, M.E., Fitzgerald, J.W., and Gosz, J.R., 1986, Sulfur processing in forest soil and litter along an elevational and vegetative gradient. Can. J. For. Res., 16:689–695.
Wehnert, M., Günther, E., and Herrmann, H., 1975, Vitamin B12-dependent methionine biosynthesis in Pseudomonas aeruginosa. Z. Allg. Mikrobiol., 15:281–286.
White, G.F., Russell, N.J., and Day, M.J., 1985, A survey of sodium dodecyl-sulfate (SDS) resistance and alkylsulfatase production in bacteria from clean and polluted river sites. Environ. Poll. Series A, 37:1–11.
Wright, D.P., Knight, CG., Parker, S.G., Christie, D.L., and Roberton, A.M., 2000, Cloning of a mucin-desulfating sulfatase gene from Prevotella strain RS2 and its expression using a Bacteroides recombinant system. J. Bacteriol., 182:3002–3007.
Zhao, Q.X. and Poole, K., 2000, A second tonB gene in Pseudomonas aeruginosa is linked to the exbB and exbD genes. FEMS Microbiol. Lett., 184:127–132.
Zürrer, D., Cook, A.M., and Leisinger, T., 1987, Microbial desulfonation of substituted naphthalenesulfonic acids and benzenesulfonic acids. Appl. Environ. Microbiol., 53:1459–1463.
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2004 Springer Science+Business Media New York
About this chapter
Cite this chapter
Kertesz, M.A. (2004). Metabolism of Sulphur-Containing Organic Compounds. In: Ramos, JL. (eds) Pseudomonas. Springer, Boston, MA. https://doi.org/10.1007/978-1-4419-9088-4_12
Download citation
DOI: https://doi.org/10.1007/978-1-4419-9088-4_12
Publisher Name: Springer, Boston, MA
Print ISBN: 978-1-4613-4789-7
Online ISBN: 978-1-4419-9088-4
eBook Packages: Springer Book Archive