Abstract
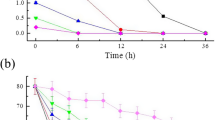
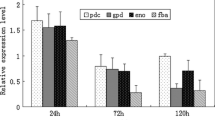
Amongst various carbon sources, xylan was found to be the sole inducer of endoxylanase production by Penicillium janthinellum MTCC 10889 in submerged cultivation. Endoxylanase synthesis by a xylan induced culture was initially repressed after a simultaneous addition of xylose, probably by the inducer exclusion mechanism, but it was resumed and achieved its highest level at a much later stage of growth (at 120 h). Xylose added after 30 h of growth cannot exert its full repressive effect. Although glucose was proved to be a more potent repressor than xylose, supplementation of salicin, an alcoholic β-glycoside containing d-glucose, with pure xylan resulted in an about 3.22 fold increase in the enzyme synthesis at 72 h followed by constant high production of the enzyme at least until the 144th h of growth. Inducing capacity of salicin in a xylan induced culture was significantly reduced when it was added after 30 h of growth. Addition of salicin and xylan help to partially overcome the repressive effect of xylose and glucose. Failure of salicin in recovering the endoxylanase synthesis in actinomycin D and cyclohexamide inhibited the xylan induced culture indicating that salicin cannot initiate the de novo synthesis of the enzyme.
Similar content being viewed by others
References
Aro, N., Pakula, T., & Penttilä, M. (2005). Transcriptional regulation of plant cell wall degradation by filamentous fungi. FEMS Microbiology Reviews, 29, 719–739. DOI: 10.1016/j.femsre.2004.11.006.
Benedetti, A. C. E. P., da Costa, E. D., Aragon, C. C., dos Santos, A. F., Goulart, A. J., Attili-Angelis, D., & Monti, R. (2013). Low-cost carbon sources for the production of a thermostable xylanase by Aspergillus niger. Revista de Ciências Farmacêuticas Básica e Aplicada, 34, 25–31.
Biely, P., & Petráková, E. (1984). Novel inducers of the xylandegrading enzyme system of Cryptococcus albidus. Journal of Bacteriology, 160, 408–412.
Biswas, S. R., Mishra, A. K., & Nanda, G. (1988). Induction of xylanase in Aspergillus ochraceus. Folia Microbiologia, 33, 355–359. DOI: 10.1007/bf02925844.
Calero-Nieto, F., Di Pietro, A., Roncero, M. I., & Hera, C. (2007). Role of the transcriptional activator XlnR of Fusarium oxysporum in regulation of xylanase genes and virulence. Molecular Plant-Microbe Interactions, 20, 977–985.
Chávez, R., Bull, P., & Eyzaguirre, J. (2006). The xylanolytic enzyme system from the genus Penicillium. Journal of Biotechnology, 123, 413–433. DOI: 10.1016/j.jbiotec.2005.12.036.
Desai, T. A., & Rao, C. V. (2010). Regulation of arabinose and xylose metabolism in Escherichia coli. Applied and Environmental Microbiology, 76, 1524–1532. DOI: 10.1128/aem.01970-09.
de Vries, R. P, & Visser, J. (2001). Aspergillus enzymes involved in degradation of plant cell wall polysaccharides. Microbiology and Molecular Biology Reviews, 65, 497–522. DOI: 10.1128/mmbr.65.4.497-522.2001.
Dhiman, S. S., Sharma, J., & Battan, B. (2008). Industrial applications and future prospects of microbial xylanases: A review. BioResources, 3, 1377–1402.
Dodd, D., & Cann, I. K. O. (2009). Enzymatic deconstruction of xylan for biofuel production. Global Change Biology Bioenergy, 1, 2–17. DOI: 10.1111/j.1757-1707.2009.01004.x.
Emami, K., Nagy, T., Fontes, C. M. G. A., Ferreira, L. M. A., & Gilbert, H. J. (2002). Evidence for temporal regulation of the two Pseudomonas cellulosa xylanases belonging to glycoside hydrolase family 11. Journal of Bacteriology, 184, 4124–4133. DOI: 10.1128/jb.184.15.4124-4133.2002.
Goyal, M., Kalra, K. L., Sareen, V. K., & Soni, G. (2008). Xylanase production with xylan rich lignocellulosic wastes by a local soil isolate of Trichoderma viride. Brazilian Journal of Microbiology, 39, 535–541. DOI: 10.1590/s1517-83822008000300025.
Herold, S., Bischof, R., Metz, B., Seiboth, B., & Kubicek, C. P. (2013). Xylanase gene transcription in Trichoderma reesei is triggered by different inducers representing different hemicellulosic pentose polymers. Eukaryotic Cell, 12, 390–398. DOI: 10.1128/ec.00182-12.
Jørgensen, H., Morkeberg, A., Krogh, K. B. R., & Olsson, L. (2004). Growth and enzyme production by three Penicillium species on monosaccharides. Journal of Biotechnology, 109, 295–299. DOI: 10.1016/j.jbiotec.2003.12.011.
Joshi, C., & Khare, S. K. (2012). Induction of xylanase in thermophilic fungi Scytalidium thermophilum and Sporotrichum thermophile. Brazilian Archives of Biology and Technology, 55, 21–27. DOI: 10.1590/s1516-89132012000100003.
Krátký, Z., & Biely, P. (1980). Inducible β-xyloside permease as a constituent of the xylan-degrading enzyme system of the yeast Cryptococcus albidus. European Journal of Biochemistry, 112, 367–373. DOI: 10.1111/j.1432-1033.1980.tb07214.x.
Khucharoenphaisan, K., Tokuyama, S., Ratanakhanokchai, K., & Kitpreechavanich, V. (2010). Induction and repression of β-xylanase of Thermomyces lanuginosus TISTR 3465. Pakistan Journal of Biological Sciences, 13, 209–215. DOI: 10.3923/pjbs.2010.209.215.
Kulkarni, N., Shendye, A., & Rao, M. (1999). Molecular and biotechnological aspects of xylanases. FEMS Microbiology Reviews, 23, 411–456. DOI: 10.1111/j.1574-6976.1999.tb00407.x.
Kundu, A., & Ray, R. R. (2011). Agrowaste utilization and production of extra cellular endoxylanase by Penicillium janthinellum MTCC 10889 in solid state fermentation. International Journal of Current Research, 3, 120–124.
Mach-Aigner, A. R., Gudynaite-Savitch, L., & Mach, R. L. (2011). l-Arabitol is the actual inducer of xylanase expression in Hypocrea jecorina (Trichoderma reesei). Applied and Environmental Microbiology, 77, 5988–5994. DOI: 10.1128/aem.05427-11.
Mandal, A., Kar, S., Das Mahapatra, P. K., Maity, C., Pati, B. R., & Mondal, K. C. (2012). Regulation of xylanase biosynthesis in Bacillus cereus BSA1. Applied Biochemistry and Biotechnology, 167, 1052–1060. DOI: 10.1007/s12010-011-9523-5.
Marui, J., Tanaka, A., Mimura, S., de Graaff, L. H., Visser, J., Kitamoto, N., Kato, M., Kobayashi, T., & Tsukagoshi, N. (2002). A transcriptional activator, AoXlnR, controls the expression of genes encoding xylanolytic enzymes in Aspergillus oryzae. Fungal Genetics and Biology, 35, 157–169. DOI: 10.1006/fgbi.2001.1321.
Miyazaki, K., Hirase, T., Kojima, Y., & Flint, H. J. (2005). Medium- to large-sized xylo-oligosaccharides are responsible for xylanase induction in Prevotella bryantii B14. Microbiology, 151, 4121–4125. DOI: 10.1099/mic.0.28270-0.
Michelin, M., Polizeli, M. L. T. M., Ruzene, D. S., Silva, D. P., Vicente, A. A., Jorge, J. A., Terenzi, H. F., & Teixeira, J. A. (2011). Xylanase and β-xylosidase production by Aspergillus ochraceus: New perspectives for the application of wheat straw autohydrolysis liquor. Applied Biochemistry and Biotechnology, 166, 336–347. DOI: 10.1007/s12010-011-9428-3.
Miller, G. L. (1959). Use of dinitrosalicylic acid reagent for determination of reducing sugar. Analytical Chemistry, 31, 426–428. DOI: 10.1021/ac60147a030.
Pal, A., & Khanum, F. (2010). Production and extraction optimization of xylanase from Aspergillus niger DFR-5 through solid-state-fermentation. Bioresource Technology, 101, 7563–7569. DOI: 10.1016/j.biortech.2010.04.033.
Puspaningsih, N. N. T., Suwanto, A., Suhartono, M. T., Achmadi, S. S., Yogiara, & Kimura, T. (2008). Cloning, sequencing and characterization of the xylan degrading enzymes from Geobacillus thermoleovorans IT-08. Jurnal ILMU DASAR, 9, 177–187.
Ren, C., Chen, T., Zhang, J., Liang, L., & Lin, Z. (2009). An evolved xylose transporter from Zymomonas mobilis enhances sugar transport in Escherichia coli. Microbial Cell Factories, 8, 66. DOI: 10.1186/1475-2859-8-66.
Seiboth, B., Herold, S., & Kubicek, C. P. (2012). Metabolic engineering of inducer formation for cellulase and hemicellulase gene expression in Trichoderma reesei. In X. Wang, J. Chen, & P. Quinn (Eds.), Reprogramming microbial metabolic pathways (Series: Subcellular biochemistry, Vol. 64, Chapter 18, pp 367–390). Dordrecht, Germany: Springer. 10.1007/978-94-007-5055-5 18.
Shulami, S., Raz-Pasteur, A., Tabachnikov, O., Gilead-Gropper, S., Shner, I., & Shoham, Y. (2011). The l-arabinan utilization system of Geobacillus stearothermophilus. Journal of Bacteriology, 193, 2838–2850. DOI: 10.1128/jb.00222-11.
Sun, J., Tian, C., Diamond, S., & Glass, N. L. (2012). Deciphering transcriptional regulatory mechanisms associated with hemicellulose degradation in Neurospora crassa. Eukaryotic Cell, 11, 482–493. DOI: 10.1128/ec.05327-11.
van Peij, N. N. M. E., Visser, J., & de Graaff, L. H. (1998). Isolation and analysis of xlnR, encoding a transcriptional activator co-ordinating xylanolytic expression in Aspergillus niger. Molecular Microbiology, 27, 131–142. DOI: 10.1046/j.1365-2958.1998.00666.x.
Zadra, I., Abt, B., Parson, W., & Haas, H. (2000). xylP promoter-based expression system and its use for antisense downregulation of the Penicillium chrysogenum nitrogen regulator NRE. Applied and Environmental Microbiology, 66, 4810–4816. DOI: 10.1128/aem.66.11.4810-4816.2000.
Zhang, J., Moilanen, U., Tang, M., & Viikari, L. (2013). The carbohydrate-binding module of xylanase from Nonomuraea flexuosa decreases its non-productive adsorption on lignin. Biotechnology for Biofuels, 6, 18. DOI: 10.1186/1754-6834-6-18.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kundu, A., Ray, R.R. Effect of salicin on induction and carbon catabolite repression of endoxylanase synthesis in Penicillium janthinellum MTCC 10889. Chem. Pap. 68, 451–456 (2014). https://doi.org/10.2478/s11696-013-0478-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.2478/s11696-013-0478-6