Abstract
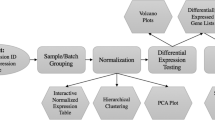
Microarrays are tools to study the expression profile of an entire genome. Technology, statistical tools and biological knowledge in general have evolved over the past ten years and it is now possible to improve analysis of previous datasets. We have developed a web interface called PHOENIX that automates the analysis of microarray data from preprocessing to the evaluation of significance through manual or automated parameterization. At each analytical step, several methods are possible for (re)analysis of data. PHOENIX evaluates a consensus score from several methods and thus determines the performance level of the best methods (even if the best performing method is not known). With an estimate of the true gene list, PHOENIX can evaluate the performance of methods or compare the results with other experiments. Each method used for differential expression analysis and performance evaluation has been implemented in the PEGASE back-end package, along with additional tools to further improve PHOENIX. Future developments will involve the addition of steps (CDF selection, geneset analysis, meta-analysis), methods (PLIER, ANOVA, Limma), benchmarks (spike-in and simulated datasets), and illustration of the results (automatically generated report).
Similar content being viewed by others
References
Baldi P., Long A.D., A Bayesian framework for the analysis of microarray expression data: regularized t -test and statistical inferences of gene changes, Bioinformatics, 2001, 17, 509–519
Kapushesky M., Kemmeren P., Culhane A.C., Durinck S., Ihmels J., Körner C., et al., Expression profiler: next generation — an online platform for analysis of microarray data, Nucleic Acids Res., 2004, 32, W465–W470
Wu C.J., Kasif S., GEMS: a web server for biclustering analysis of expression data, Nucleic Acids Res., 2005, 33, W596–W599
Montaner D., Tárraga J., Huerta-Cepas J., Burguet J., Vaquerizas J.M., Conde L., et al., Next station in microarray data analysis: Gepas, Nucleic Acids Res., 2006, W486–W491
Romualdi C., Vitulo N., Favero M.D., Lanfranchi G., Midaw: a web tool for statistical analysis of microarray data, Nucleic Acids Res., 2005, 33, W644–W649
Pochet N.L., Janssens F.A., De Smet F., Marchal K., Suykens J.A., De Moor B.L., M@cbeth: a microarray classification benchmarking tool, Bioinformatics, 2005, 21, 3185–3186
Cope L.M., Irizarry R.A., Jaffee H.A., Wu Z., Speed T.P., A benchmark for Affymetrix GeneChip expression measures, Bioinformatics, 2004, 20, 323–331
Ploner A., Miller L.D., Hall P., Bergh J., Pawitan Y., Correlation test to assess low-level processing of high-density oligonucleotide microarray data, BMC Bioinformatics, 2005, 6, 80
Harr B., Schlötterer C., Comparison of algorithms for the analysis of Affymetrix microarray data as evaluated by co-expression of genes in known operons, Nucleic Acids Res., 2006, 34, e8
Pepper S.D., Saunders E.K., Edwards L.E., Wilson C.L., Miller C.J., The utility of MAS5 expression summary and detection call algorithms, BMC Bioinformatics, 2007, 8, 273
Irizarry R.A., Bolstad B.M., Collin F., Cope L.M., Hobbs B., Speed T.P., Summaries of Affymetrix GeneChip probe level data, Nucleic Acids Res., 2003, 31, e15
Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., et al., Bioconductor: open software development for computational biology and bioinformatics, Genome Biol., 2004, 5, R80
Wu Z., Irizarry R.A., Gentleman R., Murillo F.M., Spencer F., A Model-Based Background Adjustment for Oligonucleotide Expression Arrays, J. Am. Stat. Assoc., 2004, 99, 909–917
Hubbell E., Liu W.M., Mei R., Robust estimators for expression analysis, Bioinformatics, 2002, 18, 1585–1592
Li C., Wong W.H., Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application, Genome Biol., 2001, 2, RESEARCH0032
Li C., Wong W.H. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection, Proc. Natl. Acad. Sci. U.S.A., 2001, 98, 31–36
Student, The Probable Error of a Mean, Biometrika, 1908, 1–25
Welch B.L., The significance of the difference between two means when the population variances are unequal, Biometrika, 1938, 29, 350–362
Baldi P., Long A.D. A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes, Bioinformatics 2001, 17, 509–519
Berger F., De Hertogh B., Pierre M., Gaigneaux A., Depiereux E., The “Window t-test”: a simple and powerful approach to detect differentially expressed genes in microarray datasets, Cent. Eur. J. Biol., 2008, 3, 327–344
Jain N., Thatte J., Braciale T., Ley K., O’Connell M., Lee J.K., Local-pooled-error test for identifying differentially expressed genes with a small number of replicated microarrays, Bioinformatics, 2003, 19, 1945–1951
Tusher V.G., Tibshirani R., Chu G., Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U.S.A., 2001, 98, 5116–5121, (Erratum in: Proc. Natl. Acad. Sci. U.S.A., 98, 10515)
Mann H.B., Whitney D.R., On a test of whether one of two random variables is stochastically larger than the other, Ann. Math. Stat., 1947, 18, 50–60
Breitling R., Armengaud P., Amtmann A., Herzyk P., Rank Products: A simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments, FEBS Lett., 2004, 573, 83–92
Parkinson H., Kapushesky M., Shojatalab M., Abeygunawardena N., Coulson R., Farne A., et al., ArrayExpress — a public database of microarray experiments and gene expression profiles, Nucleic Acids Res., 2007, 35, D747–D750
Edgar R., Domrachev M., Lash A.E., Gene Expression Omnibus: NCBI gene expression and hybridization array data repository, Nucleic Acids Res., 2002, 30, 207–210
Choe S.E., Boutros M., Michelson A.M., Church G.M., Halfon M.S., Preferred analysis methods for Affymetrix GeneChips revealed by a wholly defined control data set, Genome Biol., 2005, 6, R16
Bosco M.C., Puppo M., Santangelo C., Anfosso L., Pfeffer U., Fardin P., et al., Hypoxia Modifies the Transcriptome of Primary Human Monocytes: Modulation of Novel Immune-Related Genes and Identification Of CC-Chemokine Ligand 20 as a New Hypoxia-Inducible Gene, J. Immunol., 2006, 177, 1941–1955
Gaigneaux A., De Hertogh B., Berger F., Pierre M., Bareke E., Depiereux E., Discussion about ROC curves and others figures used to compare microarray statistical analyses, Proceedings of Benelux Bioinformatics Conference 2008 (BBC 2008), Maastricht, Netherlands
Affymetrix, http://www.affymetrix.com/support/technical/sample_data/datasets.affx
Author information
Authors and Affiliations
Corresponding author
Additional information
These authors contributed equally to this work
Electronic supplementary material
About this article
Cite this article
Berger, F., De Hertogh, B., Pierre, M. et al. PHOENIX, a web interface for (re)analysis of microarray data. cent.eur.j.biol. 4, 603–618 (2009). https://doi.org/10.2478/s11535-009-0055-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.2478/s11535-009-0055-8