Abstract
Background
Rapidly evolving advances in the understanding of theorized unique driver mutations within individual patient’s cancers, as well as dramatic reduction in the cost of genomic profiling, have stimulated major interest in the role of such testing in routine clinical practice. The aim of this study was to report our initial experience with genomic testing in heavily pretreated breast cancer patients.
Methods
Patients with primary or recurrent breast cancer managed at any of our five hospitals and whose malignancy had failed to respond to therapy or had progressed on all recognized standard-of-care options were offered the opportunity to have their cancer undergo next-generation sequencing genomic profiling.
Results
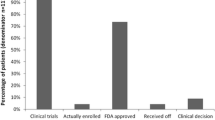
Of a total of 101 patients, 98 (97 %) had at least one specific genomic alteration identified. A total of 465 different somatic genetic abnormalities were revealed in this group of patients. Although 52 % of patients were found to have an abnormality for which an U.S. Food and Drug Administration (FDA)-approved drug was available, 69 % of patients had an FDA-approved agent for an indication other than breast cancer. The most common genomic alterations of potential clinical consequence were PIK3 (25 %), FGFR1 (16 %), AKT (11 %), PTEN (10 %), ERBB2 (8 %), JAK2 (6 %), and RAF1 (5 %).
Conclusions
Almost all advanced breast cancers possess at least one well-characterized genomic alteration that might be actionable at the clinical level. Further, in most cases, a plausible argument can be advanced for the potential biological and clinical relevance of an FDA-approved antineoplastic agent not currently indicated in the treatment of breast cancer.

Similar content being viewed by others
References
Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29.
Sabatier R, Goncalves A, Bertucci F. Personalized medicine: present and future of breast cancer management. Crit Rev Oncol Hematol. 2014;pii:S1040-8428(14)00051-1.
Mitri Z, Constantine T, O’Regan R. The HER2 receptor in breast cancer: pathophysiology, clinical use, and new advances in therapy. Chemother Res Pract. 2012;2012:743193.
Carey LA, Perou CM, Livasy CA, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;295:2492–502.
Eroles P, Bosch A, Perez-Fidalgo JA, Lluch A. Molecular biology in breast cancer: intrinsic subtypes and signaling pathways. Cancer Treat Rev. 2012;38:698–707.
Giordano A, Tagliabue E, Pupa SM. Promise and failure of targeted therapy in breast cancer. Front Biosci (Schol Ed). 2012;4:356–74.
Curtis C, Shah SP, Chin SF, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–52.
Ma CX, Ellis MJ. The Cancer Genome Atlas: clinical applications for breast cancer. Oncology (Williston Park). 2013;27:1263–9.
Cizkova M, Dujaric ME, Lehmann-Che J, et al. Outcome impact of PIK3CA mutations in HER2-positive breast cancer patients treated with trastuzumab. Br J Cancer. 2013;108:1807–9.
Berns K, Horlings HM, Hennessy BT, et al. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. 2007;12:395–402.
Nagata Y, Lan KH, Zhou X, et al. PTEN activation contributes to tumor inhibition by trastuzumab, and loss of PTEN predicts trastuzumab resistance in patients. Cancer Cell. 2004;6:117–27.
Serra V, Markman B, Scaltriti M, et al. NVP-BEZ235, a dual PI3K/mTOR inhibitor, prevents PI3K signaling and inhibits the growth of cancer cells with activating PI3K mutations. Cancer Res. 2008;68:8022–30.
Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–26.
Kean S. Breast cancer. The “other” breast cancer genes. Science. 2014;343(6178):1457–9.
Ellsworth RE, Decewicz DJ, Shriver CD, Ellsworth DL. Breast cancer in the personal genomics era. Curr Genomics. 2010;11:146–61.
Balko JM, Giltnane JM, Wang K, et al. Molecular profiling of the residual disease of triple-negative breast cancers after neoadjuvant chemotherapy identifies actionable therapeutic targets. Cancer Discov. 2014;4:232–45.
Jeselsohn R, Yelensky R, Buchwalter G, et al. Emergence of constitutively active estrogen receptor-alpha mutations in pretreated advanced estrogen receptor-positive breast cancer. Clin Cancer Res. 2014;20:1757–67.
Saal LH, Holm K, Maurer M, et al. PIK3CA mutations correlate with hormone receptors, node metastasis, and ERBB2, and are mutually exclusive with PTEN loss in human breast carcinoma. Cancer Res. 2005;65:2554–9.
Stemke-Hale K, Gonzalez-Angulo AM, Lluch A, et al. An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res. 2008;68:6084–91.
Quintas-Cardama A, Verstovsek S. Molecular pathways: Jak/STAT pathway: mutations, inhibitors, and resistance. Clin Cancer Res. 2013;19:1933–40.
Zhao W, Du Y, Ho WT, Fu X, Zhao ZJ. JAK2V617F and p53 mutations coexist in erythroleukemia and megakaryoblastic leukemic cell lines. Exp Hematol Oncol. 2012;1:15.
Gonzalez-Angulo AM, Ferrer-Lozano J, Stemke-Hale K, et al. PI3K pathway mutations and PTEN levels in primary and metastatic breast cancer. Mol Cancer Ther. 2011;10:1093–101.
Lee SY, Haq F, Kim D, et al. Comparative genomic analysis of primary and synchronous metastatic colorectal cancers. PLoS One. 2014;9:e90459.
Liedtke C, Broglio K, Moulder S, et al. Prognostic impact of discordance between triple-receptor measurements in primary and recurrent breast cancer. Ann Oncol. 2009;20:1953–8.
Von Hoff DD, Stephenson JJ Jr, Rosen P, et al. Pilot study using molecular profiling of patients’ tumors to find potential targets and select treatments for their refractory cancers. J Clin Oncol. 2010;28:4877–83.
Acknowledgment
Supported in part by Cancer Treatment Centers of America®.
Disclosure
The authors declare no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Staren, E.D., Braun, D., Tan, B. et al. Initial Experience with Genomic Profiling of Heavily Pretreated Breast Cancers. Ann Surg Oncol 21, 3216–3222 (2014). https://doi.org/10.1245/s10434-014-3925-x
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1245/s10434-014-3925-x