Abstract
Eucalyptus is the premier paper pulp, short rotation plantation species grown all over the world. Genetic improvement programs integrating molecular marker tools are in progress in many parts of the globe to increase the productivity. Whole genome sequence and expressed sequence tags (ESTs) of the eucalypts paved way for introduction of molecular genetics and breeding in this genus. Different molecular characterization approaches have been used simultaneously in eucalypts, however, microsatellites or simple sequence repeats (SSRs) with their prolific characteristics could occupy a special niche in Eucalyptus genetic improvement. Further, highly informative SSRs were used for the clonal identity, genetic fidelity and in certification of breeder’s rights. Eucalyptus genetic linkage maps generated with microsatellite loci were used successfully to identify quantitative trait loci (QTLs) for various economically important traits. Progressively more numbers of microsatellites are being linked to genes associated with adaptive and functional variations, therefore making their utility broader in genetic applications. Availability of common SSR markers across the species provides an opportunity to validate the expression of QTLs across variable genetic backgrounds and accurately compare the position of QTLs in other species. Recent evidences suggest that the presence of SSRs in micro RNAs of plant species play a role in the quantitative trait expression. Similar studies in eucalypts may provide new insights into the genetic architecture of transcript-level variations and post transcriptional gene regulation. This review on eucalypts microsatellites, highlights the availability and characteristics of genomic and eSSRs and their potential in genetic analysis of natural and breeding populations and also discusses the future prospects in population genetics and marker assisted selection.
Similar content being viewed by others
Review
Introduction
Eucalyptus is the world’s leading industrial plantation species due to its fast growth, wider adaptability and multipurpose utility. Until lately, most of the Eucalyptus plantations all over the world were destined for paper production, however, presently these plantations are looking forth to support solid wood industry including veneer production (Luo et al. [2013]). Research and development inputs on eucalypts have quadrupled the plantation productivity with the current norm of 40 m3/ha/yr (ABRAF [2013]), and unleashed possibilities up to 100 m3/ha/yr with intensive management (Evans and Turnbull [2004]). Efficient breeding and clonal deployment strategies have been employed for improved planting stock development. Efforts on molecular breeding and molecular genetic analysis are underway in eucalypts to accelerate breeding and conservation. Different kinds of DNA markers have been employed for a variety of purposes, including population genetics and marker assisted selection (MAS). One of the most frequently used DNA markers in eucalypts since 1996 are microsatellites or simple sequence repeats (SSRs) (Byrne et al. [1996]). SSRs possess characteristics such as ubiquitous distribution in genome, locus specificity, co-dominance, multi allelism, high mutation rate, heterozygous, transferability across species and associated with the gene expression and function. Hence, these markers are considered to be ideal for conservation genetics, genetic diversity assessment, variety protection, and construction of high-resolution genetic maps to link phenotypic and genotypic variation. The importance of microsatellites for plant genome analysis has been highlighted on several occasions (Varshney et al. [2005]; Kalia et al. [2011]). Earlier, genomic SSRs (gSSRs) were developed by isolating and sequencing clones containing putative SSR regions, which are costly and time consuming. Subsequently, development of online databases like GenBank led to the generation of Expressed Sequence Tag (EST) derived-SSRs (eSSRs) which are present in transcribed regions of the genome. However, in recent times, tremendous data on gSSRs and eSSRs are made available through next generation sequencing (NGS) methods (Zalapa et al. [2012]; Kudapa et al. [2014]) which can readily be used in population genetics and breeding applications.
In eucalypts, primarily genomic SSRs were developed for very few commercially important species like E. urophylla, E. grandis, E. globulus and E. nitens. However, high synteny of genome existing across eucalypt species has benefited many DNA data deficient eucalypts taxa, thus excellent SSR transferability was witnessed across species (da Silva et al. [2009]; Acuna et al. [2012a]). SSRs were used in eucalypts for multiple purposes such as species identification, phylogeny, hybrid authenticity, genetic diversity studies, genetic mapping and Quantitative trait loci (QTL) localization (Myburg et al. [2007]; Grattapaglia et al. [2012]). SSRs also offer a much higher level of automation which is crucial to manage with the great number of individuals generally handled in eucalypt breeding. These markers have advantages over high throughput SNP markers, because the majority of SNPs is bi-allelic and information (heterozygosity) generated is low, whereas SSR mutational rates are a lot higher and the DNA slippage process creates a number of new alleles leading to the generation of maximal information (Ellegren [2004]). Many studies on the comparison between SSRs and SNPs have proven that large numbers of SNP loci were required to replace highly polymorphic SSRs in studies of diversity and relatedness (Hamblin et al. [2007]; Yang et al. [2011]). Further, SSR markers were always preferred as framework markers for developing consensus linkage map, composite integrated linkage map and comparative map between the species (Garcia et al. [2011]; Hudson et al. [2012]).
Microsatellite resources in eucalypts
Based on the published literature, till date, a total of 505 genomic SSRs (gSSRs) and 758 validated EST-SSRs (eSSRs), 35 chloroplast SSRs (cpSSRS) and 8 gene based SSRs (CG-SS1Rs) have been applied in different species of Eucalyptus. The details on SSR marker code, source species and number of SSRs developed are given in the Table 1. A largest collection of both gSSRs and eSSRs (~300 SSRs) was developed from E. grandis and E. urophylla with the prefix as Embra (Brondani et al. [1998], [2002], [2006]; Faria et al. [2010], [2011]). Other major resources include the SSRs with the prefix Emcrc (40 SSRs) developed from E. globulus (12 loci; Steane et al. [2001]), Corymbia variegata (14 loci; Jones et al. [2001]) and Corymbia citriodora subsp variegata (14 loci; Shepherd et al. [2006]). The SSRs with prefixes En, Es, Eg and El were developed from species such as E. nitens (8 SSRs), E. sieberi (8 SSRs), E. globulus (26 SSRs) and E. leucoxylon (13 SSRs) respectively (Byrne et al. [1996]; Glaubitz et al. [2001]; Ottewell et al. [2005]). A set of 35 chloroplast DNA microsatellites was developed based on the full cp-DNA sequence of E. globulus (Steane et al. [2005]). The ISSR-enrichment technique was exploited for the development of five SSR loci in E. grandis (Van der Nest et al. [2000]). Very few SSR loci were developed from E. urophylla and E. pilularis (Payn et al. [2008]; Sexton et al. [2010]). NGS methods such as 454 sequencing was applied to isolate ten SSRs from E. victrix (Nevill et al. [2013]). A recent study in Eucalyptus found that microsatellites occupy approximately 0.6 percent of the overall genome (Ranade et al. [2014]).
Whole-genome sequencing of E. camaldulensis and E. grandis have been completed by the Kazusa DNA Research Institute, Japan and DOE Joint Genome Institute (JGI), USA in collaboration with members of the Eucalyptus Genome Network (EUCAGEN) respectively (Hirakawa et al. [2011]; Myburg et al. [2014]). Additionally, transcriptome resources were generated from various tissues including xylem, phloem, root, shoot, leaf and reproductive tissues from species such as E. grandis, E. gunnii, E. globulus, E. camaldulensis and E. tereticornis (Mizrachi et al. [2010]; http://web.up.ac.za/eucagen/; http://eucgenie.org/; Healey et al. [2014]). All these transcriptome resources have promoted the development of SSR markers in silico and many of which were used for diverse purposes across eucalypt species (Ceresini et al. [2005]; Rabello et al. [2005]; Yasodha et al. [2008]; Rengel et al. [2009]; He et al. [2012]; Zhou et al. [2014]). Recently, gene specific microsatellites were developed from E. grandis, E. globulus and E. gomphocephala (Acuna et al. [2012b]; Bradbury et al. [2013a]). Table 2 provides the list of EST-SSRs developed in various species of eucalypts.
The frequency of occurrence of microsatellites had varied in different databases of Eucalyptus for example, 12.9% in NCBI database (Yasodha et al. [2008]) 13.3% in EUCAWOOD (Rengel et al. [2009]), 25.5% and 29% in FORESTs database (Rabello et al. [2005]; Ceresini et al. [2005]). The type of SSRs found in the ESTs varied among the transcriptome analyzed. In general, amongst the SSR motifs, the dimeric and trimeric were most abundant followed by other types. This is in consonance with many other plant species belonging to monocots as well as dicots. The most represented di-nucleotide was AG/TC (72.5%) motif followed by the trimeric CCG/GGC, AAG/TTC, and AGA/TCT (12.81%) (Ceresini et al. [2005]; Rabello et al. [2005]; Yasodha et al. [2008]; Rengel et al. [2009]; Ranade et al. [2014]). The details on different types of motifs are shown in Table 3. These motifs have also been found to be predominant dinucleotide repeats and trinucleotide repeats respectively in many plant species (Zhou et al. [2014]). The BAC clones of eucalypts had comparatively less numbers of SSR frequency (Paiva et al. [2011]). Most of the SSRs isolated from genomic libraries were targeted for simple dinucleotide repeats, particularly AG/TC motifs except a few had compound, interrupted and trinucleotide motifs (Glaubitz et al. [2001]; Ottewell et al. [2005]; Brondani et al. [2006]). Unique in silico methods were developed to extract and transfer the highly conserved orthologus genic SSR regions from E. globulus to E. camaldulensis, a species with less genomic information, and such novel SSRs were useful for parentage analysis, confirmation of interspecific hybrid and genotyping of seedling seed orchard (Nagabhushana et al. [2011]). A study to mine SSRs in silico from 22298 EST sequences of eucalypts revealed that primers could be designed for 1244 microsatellites, of which 182 were selected for characterization based on polymorphism status among species (Grattapaglia et al. [2014]).
Cross-species transferability
Microsatellite markers are generally transferable across related genera and the genetic distance among the species limits the percent transferability. The cross species transferability provides a potential source of codominant markers for many related species and facilitates evolutionary, ecological, and conservation studies across the species. Since the display of cross-species transferability of microsatellite markers is high in eucalypts, attempts have been made to identify a useful number of primer sets of high utility in a wide range of species. Details of the SSR source species and the species in which the SSRs cross amplified are given in the Table 4. The cross species transferability of genomic SSR markers was comparatively higher among the species of Eucalyptus and the transferability rate dropped down in the species of Corymbia and Angophora (Steane et al. [2001]). Initially, Byrne et al. ([1996]) tested transferability of four nuclear microsatellite markers from E. nitens and found that they were transferable (50%) to sub genera Symphyomyrtus and Monocalyptus but not to the genus Corymbia. However, modifications in the PCR amplification protocols could improve the transfer rate across genera (Shepherd et al. [2006]). Many of the SSR loci isolated from E. grandis and E. urophylla were cross amplified in different species like E. globulus, E. nitens, E. pilularis, E. urophylla, E. pyrocarpa, E. camaldulensis and E. tereticornis (Bundock et al. [2000]; Glaubitz et al. [2001]; Steane et al. [2001]; Agrama et al. [2002]; Ottewell et al. [2005]; Arumugasundaram et al. [2011]; Subashini et al. [2013]). Similarly, the eSSRs were successfully cross amplified in several species like E. grandis, E. saligna, E. dunnii, E. viminalis, E. camaldulensis, E. urophylla and E. tereticornis (Neves et al. [2011]; Faria et al. [2010], [2011]; Acuna et al. [2012b]; Hudson et al. [2012]; Petroli et al. [2012]; Breed et al. [2012]; He et al. [2012]; Bradbury et al. [2013a], [b]; Bradbury and Krauss [2013]). Recently, gene-homologous eSSRs designed for E. gomphocephala were transferred to E. marginata, E. camaldulensis, and E. victrix (Bradbury et al. [2013a]).
Genotyping of SSRs and allele variations in eucalypts
Genotyping studies with microsatellites generally preferred dinucleotide repeats because they bring off high polymorphism and display more variation among individuals. In certain occasions the dinucleotide repeats based allelic variation results in shadow bands or stutter bands during electrophoresis thus leading to genotyping errors (Hoffman and Amos [2005]). Hence, the genotyping applications requiring high precision like clonal certification, microsatellite markers with tetra and penta nucleotide motifs were recommended (Faria et al. [2010]). Loci with higher length repeats provided an advantage of accurate allele calling due to their larger allele size difference. In eucalypts, presence of tetra, penta and hexanucleotide allowed easy allele calling which was challenging when di and trinucleotide motif SSRs were used (Faria et al. [2010]). Presence of null allele, i.e., a microsatellite locus that consistently fails to amplify to detected levels via the polymerase chain reaction (PCR) is not uncommon in eucalypts (Glaubitz et al. [2001]). In general, microsatellite null alleles at low frequencies are unlikely to introduce serious biases into population genetic analysis (Dakin and Avise [2004]). Nevertheless, microsatellite null alleles can cause egregious errors when they are used for genetic mapping experiments at family level. There are methods and software packages available to handle the null alleles and heterozygote deficiency in various genetic analyses (Chapuis and Estoup [2007]; Chybicki and Burczyk [2009]). In a genetic mapping study, it was found that 20 out of 241 segregating SSR loci were observed to have null alleles (Brondani et al. [2006]). Increased occurrence of null alleles have been observed when attempting to transfer microsatellites across related species (Faria et al. [2010]; Bradbury et al. [2013a], [b]; Bradbury and Krauss [2013]) and appropriate strategies need to be used in handling such data. Genotyping with microsatellites markers on large scale using DNA sequencing instruments demonstrated very high accuracy of allele sizing and binning to avoid the scoring errors. Although various methods of primer labeling and SSR allele detection in eucalypts was reported (Ottewell et al. [2005]; Missiaggia and Grattapaglia [2006]; Faria et al. [2011]; Subashini et al. [2013]), difficulties in accuracy of allele sizing continue to exist.
Allele sizes generated by the microsatellites loci have larger implications on genotyping of the individuals. Allele sizes generated by the eucalypt microsatellite loci were highly varying, as low as 50 bp was detected in E. camaldulensis (da Silva et al. [2009]). Very high levels of allelic variability were observed at different levels of populations. A pioneering study on analysis of 15 SSR loci in 32 F1 individuals of E. grandis × E. urophylla generated 9 to 26 alleles per locus with an average of 16.3 × 4.8 (Brondani et al. [1998]). Similarly, a breeding population of E. grandis with 192 selected individuals produced a total of 119 alleles with 6 SSR loci, yielding a minimum of 6 (Embra11) and a maximum of 33 alleles (Embra16), with an average of 19.8 × 9.2 alleles per locus (Kirst et al. [2005a]).
Compared to gSSRs, eSSR loci were known for low levels of polymorphism, Faria et al. ([2010]) analyzed 10 eSSRs in 6 eucalypt species wherein the number of alleles were in the range of 7–15 (E. grandis), 5–12 (E. globulus), 4–10 (E. urophylla), 6–14 (E. camaldulensis), 5–9 (E. dunnii) and 4–14 (E. saligna). Allele size variations favored multiplexing of fluorescent based automated DNA genotyping applications, wherein upto 18 loci were analyzed in a single run with 5 dye format (Faria et al. [2010]).
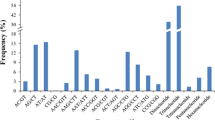
Eucalyptus microsatellites are usually highly informative as revealed through the statistics of polymorphic information content (PIC) and heterozygosity (He) (Table 5). The most widely used SSRs such as Embra, generated maximum heterozygosity value of 0.95 (Holman et al. [2003]; Jones et al. [2008]), Emcrc markers showed highest heterozygosity of 0.92, Es, En and El series loci produced 0.95, 0.91 and 0.93 (Byrne and Hines [2004]; Glaubitz et al. [2001]; Ottewell et al. [2005]). Accordingly the PIC values were also very high (0.933) in most of the studies so far reported (Kirst et al. [2004]). Because of these characteristics of microsatellites, they were used for various purposes in plant genome analysis (Figure 1).
Applications of microsatellite markers in eucalypts
The stupendous and multifaceted applications of microsatellites in tree genome analysis are shown in the Figure 1. The ubiquitous nature of microsatellites in tree genome (present in nuclear, EST, mitochondria and chloroplast genome sequences) make them the most suitable DNA markers for analysis of population genetics, phylogeny and species evolution, studies on conservation and ecology and marker assisted selection/breeding (Porth and El-Kassaby [2014]). Highly informative SSR markers generate multiple alleles, thus favoring germplasm/clonal identification, genetic integrity determination during propagation and controlled breeding, seed orchard diversity, mating system and outcrossing rate analysis (Falahati-Anbaran et al. [2007]). The multilocus nature and even distribution of microsatellites over the plant genome benefit the establishment of taxonomic identity of species and phylogeny reconstruction (Ochieng et al. [2007]). Microsatellites are ideal for conservation genetics and genetic resource management because of their selectively neutral characteristics (van Zonneveld et al. [2014]) and evolutionary processes of interest to conservation geneticists (Wang et al. [2009]).
Characterization of germplasm and individual identification
Microsatellites information on genotype is essential for effective utilization of germplasm accessions for genetic improvement for pulp yield, adventitious rooting, frost and salt tolerance, resistance to pests and diseases, efforts have continuously been made to characterize and evaluate germplasm accessions. Large amount of such germplasm and clonal accessions are maintained in tree breeding programs and clonal deployment activities of eucalypts. The high degree of polymorphism and the clear and simple co-dominant Mendelian inheritance of the SSRs has proven to be an extremely powerful system for the unique identification of Eucalyptus individuals for fingerprinting purposes and parentage testing (Kirst et al. [2005b]). Accordingly, SSR markers have been widely used for characterization of germplasm resources including clone fingerprinting, hybrid validation in controlled crosses, inter-individual genetic distance estimation, species distinction, and assignment of hybrid individuals to their most likely parent species (Stokoe et al. [2001]; Smith et al. [2003]; Grattapaglia et al. [2004]a; Kirst et al. [2005b]; Ochieng et al. [2007]; Grosser et al. [2008]; Jones et al. [2008]; Payn et al. [2008]; Rao et al. [2008]; Sampson and Byrne [2008]; Butcher et al. [2009]; Barbour et al. [2010]; Faria et al. [2010]; Shepherd et al. [2010]; Ribeiro et al. [2011]; Arumugasundaram et al. [2011]; Subashini et al. [2013]; Wheeler et al. [2013]) Clonal fingerprinting generally requires larger allele size differences for multiplexing, precise and stable allele sizing for deployment across laboratories. Hence, SSR markers with high motif length provided possibilities for greater utilization towards individual identification (Faria et al. [2011]). SSR markers play an important role in designing breeding populations and function as decision support tool in genetic conservation programs. Use of SSR markers in germplasm characterization of Jatropha curcus showed that the germplasm has limited diversity and the necessity of additional collections for species improvement (Alves et al. [2011]). Similarly, SSRs are the most sought after DNA markers for germplasm characterization in several tree species like avocado (Gross-German and Viruel [2013]), Psidium guajava (Sitther et al. [2014]), Olive (Trujillo et al. [2014]), Tamarix (Terzoli et al. [2014]) and Prunus (Khadivi-Khub et al. [2014]).
Parentage testing and gene flow studies
Seed orchards of eucalypt species are established with individuals having high genetic diversity and low levels of inbreeding to achieve the high genetic gain in progenies. Microsatellite markers played a major role in attaining these goals by displaying mating system, degree of contamination, variation in outcrossing rates, gene flow pattern and paternal contribution in the seed orchard. The Table 6 provides the information of SSR loci used for parentage analysis and gene flow studies. Parentage assignments based on microsatellite allele information could lead to the identification of appropriate parents contributing for potential progeny (Grattapaglia et al. [2004]a). Microsatellite diversity was considered for the designing of seed orchard in E. dunnii (Zelener et al. [2005]) and E. globulus (Dini et al. [2011]) thus reducing the risks of inbreeding. Eucalyptus has predominantly outcrossing breeding system, nevertheless the rate of outcrossing varies among the species and populations. E. grandis seed orchard in exotic conditions showed the maximum outcrossing rate of 96.7% however, the pollen contamination in the seed orchard was 39.2% (Chaix et al. [2003]). Similarly, another study on E. grandis seed orchard in Brazil analyzed with 14 SSR markers had 8.3% selfing and 29% pollen contamination (Grattapaglia [2004]b). In native locations, E. grandis seed orchard populations showed a selfing rate of 14% and the paternity analysis showed that 46% seeds were due to contamination (Jones et al. [2008]). On contrary, a well isolated clonal seed orchard of E. nitens had an outcrossing rate of 0.85% with relatively low pollen contamination of 4.5% (Grosser et al. [2008]). A small breeding arboretum of E. globulus in Australia was analyzed with four highly polymorphic SSRs revealed 47.9% outcrosssing rate and 17.6% contamination rate (Rao et al. [2008]). These results indicate that microsatellites are the best tools for predicting the seed orchard performance and revealing the importance of strategies to arrest outside pollen like flowering asynchrony and isolation distance of adjacent populations. Recently, Correia et al. ([2011]) showed that tetra-, penta- and hexa nucleotide microsatellites were more informative than the di and tri nucleotide markers and specific SNPs employed for assessment of parentage and individual identification. Thus, construction of a panel of markers to assess parentage would allow considerable inputs for designing of seed orchards consequently increased productivity from half-sib eucalypt populations.
The natural populations of eucalypts were subjected to microsatellite based genetic analysis for gene flow estimation and population differentiation. The E. camaldulensis species complex in its natural range was assessed for its genetic structure with 15 microsatellite loci and concluded that the populations belongs to several subspecies with intergrade zones and breeding programs should not treat the species as a single genetic entity (Butcher et al. [2009]). Similarly, E. globulus species complex was surveyed with 9 microsatellite loci displayed the existence of intergrade populations and the possibilities of 4 subspecies were identified (Jones et al. [2013]). The presence of spatial genetic differentiation and large breeding zones in E. globulus populations in its native range revealed a distant pollen movement (Steane et al. [2006]; Yeoh et al. [2012]). Whereas, in E. urophylla populations of Indonesian islands, low levels of genetic differentiation across populations with high levels of gene diversity within populations were recorded (Payn et al. [2008]). Such information will have large implications in breeding of these economically important plantation species.
Eucalyptus species with regional distribution were analyzed for their pollen flow and pattern of genetic diversity. These studies involve microsatellite allele diversity largely indicated that in remnant and fragmented populations of E. curtisii (Smith et al. [2003]), E. leucoxylon (Ottewell et al. [2005]), E. wandoo (Byrne et al. [2008]), E. benthamii (Butcher et al. [2005]), E. gomphocephala (Bradbury et al. [2013a], [b]; Bradbury and Krauss [2013]), E. incrassata (Breed et al. [2012]) and E. loxophleba (Sampson and Byrne [2008]) the genetic differentiation were high across the locations and pollen based gene flow is well maintained. On the other hand, fragmentation led to high degree of clonality and inbreeding in few of the species (Smith et al. [2003]; Butcher et al. [2005]). Further, pollen being the main mode of gene flow, genetic differentiation in quantitative traits was maintained by natural selection (Bloomfield et al. [2011]). Nuclear and chloroplast microsatellite markers were deployed to examine the spatial distribution of genetic diversity in E. pauciflora in Tasmania and the results revealed the route of seed and pollen dispersal and population migration pattern (Gauli et al. [2014]). Effective pollen movement and spatial proximity of different species of eucalypts, which has limited reproduction barriers provides sufficient information for distances required for buffer zone in seed orchards and to maintain genetic integrity of breeding populations in exotic and natural locations. These distinctive features revealed by the microsatellite markers have a high significance in sourcing of seeds and designing breeding and conservation programs (Bacles et al. [2009]; Barbour et al. [2010]; Shepherd and Raymond [2010]).
Linkage map and QTL identification
Microsatellite markers are regarded to be the tools in marker assisted selection and they are widely utilized for genetic mapping in many forest trees and they are extremely suitable for QTL localization and comparative mapping purposes. Detailed information on the genetic maps developed till date was reviewed by Grattapaglia et al. ([2012]). Most of the genetic mapping studies in eucalypts targeted the commercially important species such as E. grandis, E. urophylla, E. grandis, E. globulus, E. camaldulensis and E. nitens (Bundock et al. [2000]). The Additional file 1: Table S1 shows the SSR markers mapped in genetic linkage maps of different species. In eucalypt inter-specific hybrids, the first genetic map was developed with dominant RAPD markers using pseudotestcross approach (Grattapaglia and Sederoff [1994]). Immediately after the development of genetic maps, QTL localization for growth, adventitious rooting and wood properties were carried out rapidly. Microsatellite linkage mapping was possible only in 1998 for the Eucalyptus grandis × E. urophylla interspecific cross with 20 highly informative Embra SSR loci (Brondani et al. [1998]). Congruity of genetic linkage maps of different eucalypt species had an advantage of consolidating linkage groups across species and quantitative trait loci influencing the traits of interest. The marker correspondence across linkage grouping and position of the SSR loci across genetic maps of eucalypt species was almost similar with few exceptions (See Additional file 1 for linkage group information). The first QTL study involving SSR loci in Eucalyptus for a series of wood properties was reported by Thamarus et al. ([2002]). Vegetative propagation traits were located on homeologous linkage groups of E. grandis, E. urophylla, E. tereticornis and E. globulus (Marques et al. [2002]). Further, integrated linkage maps with different types of DNA markers were developed for many species of eucalypts. The pure species genetic map for E. camaldulensis was developed with RAPDs, RFLPs and SSRs by selecting highly divergent parent trees for mapping population generation (Agrama et al. [2002]). With the development of new SSR markers, Brondani et al. ([2006]) could generate a comprehensive consensus linkage map by including SSR loci information from various eucalypts species. In the recent years, along with the next generation markers like SNPs, SFPs and DArT markers, SSRs are used as framework markers to confirm the linkage groups and position of markers. The SSR markers were used to estimate linkage disequilibrium in eucalypts but reported to be lasting very fast with every 200 bp approximately (Arumugasundaram et al. [2011]). However, in several instances SSR loci were found to be in close association with quantitative traits. For example, in E. grandis breeding population Embra125 and Embra1071 were found to be in linkage equilibrium with rust resistance loci Ppr 1 at 9.5 and 7 recombination, respectively (Mamani et al. [2010]). Embra125 loci was found to be closely linked with rust resistance in E. grandis [(E. grandis) × (E. urophylla × E. grandis)] explaining 42% of the phenotypic variation (Rosado et al. [2010]). Recently, between eSSR markers Embra1656 and Embra1071 (16.4 and 1.4 cM away, respectively) the QTLs governing moderate proportion of the genetic variation (11.5%) for rust resistance was identified (Alves et al. [2011]).
In E. globulus putative QTL for Mycosphaerella cryptica resistance was closely associated with microsatellite marker Embra48 (Freeman et al. [2008b]). Similarly, Embra12 was in closely linked to foliar concentrations of terpenes and formylated phloroglucinol compounds in E. nitens and E. globulus (Henery et al. [2007]; Freeman et al. [2008a]). Embra173 alone explained 53.8% variation for formylated phloroglucinols in the foliage of E. globulus (Freeman et al. [2008b]). In another study by Freeman et al. ([2009]) in E. globulus several Embra SSR loci were linked to wood properties and growth traits. Additive and dominant QTLs were found in tight linkage with SSR markers tested for drought tolerance in E. grandis × E. urophylla hybrid clones (Teixeira et al. [2011]).
The presence of generic genomic regions was validated through SSR markers, which enabled the identification of orthologous QTL regions for wood properties in E. nitens and E. globulus (Thumma et al. [2010]) and E. urophylla and E. grandis (Gion et al. [2011]). Genic SSRs were found to be largely associated with economically important traits in many plant species. Eucalyptus transcriptome sequencing projects led to the development of many eSSR loci and many were used in genetic maps (Faria et al. [2011]). Eucalyptus species have valuable SSR resources for comparative genomic studies and they also serve as framework markers for construction of a consensus map across species. Addition of fully informative microsatellites on the framework map permits linkage map homology, QTL and candidate gene positions across different eucalypt species (Freeman et al. [2009]). Eucalyptus SSRs were proved again for their colinearity across different species and linkage to physical correspondence on the reference genome sequence (Grattapaglia et al. [2014]).
Future prospects
Microsatellites have major roles to play in various spheres of eucalypts genetics and improvement. They are one of the three major classes of genetic variations along with SNPs (single nucleotide polymorphisms) and CNVs (copy number variations) and have many important biological functions (Gemayel et al. [2010]). Recent evidences suggest that variations in microsatellites may lead to phenotypic changes (Joy and Soniya [2012]) and adaptive evolution (Fidalgo et al. [2006]). The available genome sequence of the eucalypts genome does not diminish the importance of microsatellites, as these markers will extend annotated genome resources of sequenced Eucalyptus species to genetic study/breeding in different eucalypt species.
Genomic SSRs have a broad range of applications, and in particular being neutral markers, which are not linked to any particular trait, but most probably offer a representation of the underlying genetic diversity in wild populations and to target populations for conservation (van Zonneveld et al. [2012]). Further, microsatellites enable the rapid identification of cryptic species and have been used successfully to identify species hybrids in many tree species including eucalypts. They contribute to a better understanding of the processes involved in the development of contemporary patterns of variation, including the regional contraction and expansion of populations and refugia (Nevill et al. [2014]).
Several reports confirmed the presence of SSRs in transcription factors and promoters of genes for facilitating transcriptional plasticity. Hence, identification of new gSSRs and eSSRs would pave way for better understanding of the Eucalyptus genome. Genic SSR markers could represent the new class of functional markers, finding use in evolutionary studies, comparative mapping, candidate gene association mapping, gene discovery and molecular breeding (Shi et al. [2014]). In silico SSR polymorphism analysis, a novel access to selecting polymorphic markers is currently advocated to reduce the cost and to increase the efficiency of SSR development.
Further, enough care should be exercised while handling null alleles, imperfect repeats, and allelic dropout, equally they can lead to an overestimation of observable alleles, a decrease in observed heterozygosity, and an increase in the apparent level of inbreeding. On the basis of microsatellite analysis, spatial genetic structure (SGS) can be estimated to delineate provenances of eucalypts. In addition, non-denaturing FISH (ND-FISH) can be used to compare the distribution of SSRs to determine whether the range of molecular diversity shown by these highly polymorphic sequences is reflected at the chromosome level. They are also ideally used as anchor markers in molecular linkage maps and in generation of consensus maps across species and that can be highly saturated with DArT and SNP/genotyping by sequencing markers (GBS). Development of a high-density consensus genetic map with SSRs in an important chromosomal interval provides eucalypt molecular breeding programs with a better choice regarding the quality of markers and a higher probability of polymorphic markers. They play a major role in aligning linkage map due to their high transferability and have a functional role in trait variation and to see the conservation and diversification of gene order across the species of eucalypts.
Conclusions
Microsatellite markers play a major role in eucalypts at different levels of genetic improvement. The inherent potentiality of these marks to distinguish closely related individuals is increasingly encouraging for the mining of more and more SSRs for placing on linkage groups and other genetic studies. Microsatellites containing DNA sequences and their functional role in the eucalypt genome were investigated and detected linkage-to-physical position for a large number of microsatellites. Recently, two hundred and twenty three new microsatellite markers were surveyed for allelic diversity and added to the existing eucalypt SSR map, bringing the total number of genetically mapped SSR loci to >400 and strengthening the comparative genome mapping (Grattapaglia et al. [2014]).
Additionally, the miRNA-SSR markers, i.e., presence of SSRs in precursors of miRNA candidates (Joy et al. [2013]) brought a new biological significance to microsatellites, wherein the microRNAs (miRNAs) play a major role in post transcriptional gene silencing. The length variation of the SSRs in salt responsive miRNA genes provided sensitivity to salinity adaptation of Oryza sativa (Mondal and Ganie [2014]). Any prosperous utilization in tree breeding, SSRs is required in greater numbers. Therefore, recognition of such miRNA-SSRs in eucalypts would lead to better understanding of their role in post-transcriptional gene regulations and phenotypic variations.
Additional file
References
Statistical yearbook. Brazilian Association of Forest Plantation Producers, Brasilia DF; 2013.
Acuna C, Villalba P, Garcia M, Hopp E, Marcucci S: Functional markers development and genetic diversity analysis in Eucalyptus globulus with emphasis in wood quality candidate genes. BMC Proc 2011, 5(l 7):154.
Acuna CV, Fernandez P, Villalba PV, García MN, Hopp E, Poltri SNM: Discovery, validation, and in silico functional characterization of EST-SSR markers in Eucalyptus globulus . Tree Genet Genomes 2012, 8: 289–301.
Acuna CV, Villalba PV, Garcia M, Pathauer P, Hopp HE, Marcucci Poltri SN: Microsatellite markers in candidate genes for wood properties and its application in functional diversity assessment in Eucalyptus globulus . Electron J Biotechnol 2012, 15(2):1–17.
Agrama H, George TL, Salah SF: Construction of genome map for Eucalyptus camaldulensis DEHN. Silvae Genet 2002, 51(5–6):201–206.
Alves AA, Rosado CCG, Faria DA, Da Silva Guimaraes LM, Lau D, Brommonschenkel SH, Alfenas AC: Genetic mapping provides evidence for the role of additive and non-additive QTLs in the response of inter-specific hybrids of Eucalyptus to Puccinia psidii rust infection. Euphytica 2011, 183(1):27–38.
Arumugasundaram S, Ghosh M, Veerasamy S, Ramasamy Y: Species discrimination, population structure and linkage disequilibrium in Eucalyptus camaldulensis and Eucalyptus tereticornis using SSR markers. Plos One 2011, 6(12):e28252.
Bacles CF, Brooks J, Lee DJ, Schenk PM, Lowe AJ, Kremer A: Reproductive biology of Corymbia citriodora subsp. variegata and effective pollination across its native range in Queensland, Australia. Southern Forests: J Forest Sci 2009, 71(2):125–132.
Barbour RC, Wise SL, McKinnon GE, Vaillancourt RE, Williamson GJ, Potts BM: The potential for gene flow from exotic eucalypt plantations into Australia’s rare native eucalypts. For Ecol Manag 2010, 260(12):2079–2208.
Bartholome J, Salmon F, Vigneron P, Bouvet JM, Plomion C, Gion JM: Plasticity of primary and secondary growth dynamics in Eucalyptus hybrids: a quantitative genetics and QTL mapping perspective. BMC Plant Biol 2013, 13(1):1–14.
Bloomfield JA, Nevill P, Potts BM, Vaillancourt RE, Steane DA: Molecular genetic variation in a widespread forest tree species Eucalyptus obliqua Myrtaceae on the island of Tasmania. Aust J Bot 2011, 59(3):226–237.
Bradbury D, Krauss SL: Limited impact of fragmentation and disturbance on the mating system of tuart Eucalyptus gomphocephala , Myrtaceae: implications for seed- source quality in ecological restoration. Aust J Bot 2013, 61(2):148–160.
Bradbury D, Smithson A, Krauss SL (2013a) Development and testing of new gene- homologous EST-SSRs for Eucalyptus gomphocephala Myrtaceae. Appl Plant Sci 1(8). , [http://dx.doi.org/10.3732/apps.1300004] Bradbury D, Smithson A, Krauss SL (2013a) Development and testing of new gene- homologous EST-SSRs for Eucalyptus gomphocephala Myrtaceae. Appl Plant Sci 1(8).
Bradbury D, Smithson A, Krauss SL: Signatures of diversifying selection at EST-SSR loci and association with climate in natural Eucalyptus populations. Mol Ecol 2013, 22(20):5112–5129.
Breed MF, Ottewell KM, Gardner MG, Marklund MHK, Stead MG, Harris JBC, Lowe AJ (2012) Mating system and early viability resistance to habitat fragmentation in a bird- pollinated eucalypt. Heredity. doi:10.1038/hdy.2012.72 Breed MF, Ottewell KM, Gardner MG, Marklund MHK, Stead MG, Harris JBC, Lowe AJ (2012) Mating system and early viability resistance to habitat fragmentation in a bird- pollinated eucalypt. Heredity. doi:10.1038/hdy.2012.72
Brondani RPV, Brondani C, Tarchini R, Grattapaglia D: Development, characterization and mapping of microsatellite markers in Eucalyptus grandis and E. urophylla . Theor Appl Genet 1998, 97(5–6):816–827.
Brondani RPV, Brondani C, Grattapaglia D: Towards a genus-wide reference linkage map for Eucalyptus based exclusively on highly informative microsatellite markers. Mol Genet Genom 2002, 267(3):338–347.
Brondani RPV, Williams ER, Brondani C, Grattapaglia D: A microsatellite-based consensus linkage map for species of Eucalyptus and a novel set of 230 microsatellite markers for the genus. BMC Plant Biol 2006, 6(1):20.
Bundock PC, Hayden M, Vaillancourt RE: Linkage maps of Eucalyptus globulus using RAPD and microsatellite markers. Silvae Genet 2000, 49: 223–232.
Butcher PA, Skinner AK, Gardiner CA: Increased inbreeding and inter-species gene flow in remnant populations of the rare Eucalyptus benthamii . Conserv Genet 2005, 6(2):213–226.
Butcher PA, Mc Donald MW, Bell JC: Congruence between environmental parameters, morphology and genetic structure in Australia’s most widely distributed eucalypt, Eucalyptus camaldulensis . Tree Genet Genomes 2009, 5(1):189–210.
Byrne M, Hines B: Phylogeographical analysis of cpDNA variation in Eucalyptus loxophleba Myrtaceae. Aust J Bot 2004, 52(4):459–470.
Byrne M, Garcia MMI, Uren T, Smith DS, Moran GF: Conservation and genetic diversity of microsatellite loci in the genus Eucalyptus . Austr J Bot 1996, 44: 331–341.
Byrne M, Elliott CP, Yates CJ, Coates DJ: Maintenance of high pollen dispersal in Eucalyptus wandoo , a dominant tree of the fragmented agricultural region in Western Australia. Conserv Genet 2008, 9(1):97–105.
Ceresini PC, Silva PCLS, Missio RF, Souza EC, Fischer CN, Guillherme IR, Gregorio I, Da Silva EHT, Cicarelli RMB, Da Silva MTA: Satellyptus: analysis and database of microsatellites from ESTs of Eucalyptus . Genet Mol Biol 2005, 28(3):589–600.
Chaix G, Gerber S, Razafimaharo V, Vigneron P, Verhaegen D, Hamon S: Gene flow estimation with microsatellites in a Malagasy seed orchard of Eucalyptus grandis . Theor Appl Genet 2003, 107(4):705–712.
Chapuis MP, Estoup A: Microsatellite null alleles and estimation of population differentiation. Mol Bio Evol 2007, 24(3):621–631.
Chybicki IJ, Burczyk J: Simultaneous estimation of null alleles and inbreeding coefficients. J Hered 2009, 100(1):106–113.
Correia L, Faria D, Grattapaglia D: Comparative assessment of SNPs and microsatellites for fingerprinting, parentage and assignment testing in species of Eucalyptus . BMC Proc 2011, 5: 41.
da Silva JM, De Sousa ACB, De Souza AP, Mori ES, Freitas MLM, Sebbenn AM, De Moraes MLT: Development and characterization of 14 microsatellite loci from an enriched genomic library of Eucalyptus camaldulensis Dehnh. Conserv Genet Resou 2009, 1(1):465–469.
Dakin EE, Avise JC: Microsatellite null alleles in parentage analysis. Heredity 2004, 93(5):504–509.
Dini TD, Bennadji Z, Cabrera M, Centurion C, Resquin F, Balmelli G: Use of SSR- tools for clone certification in Uruguayan Eucalyptus grandis and Eucalyptus dunnii breeding programs. BMC Proc 2011, 5: 58.
Ellegren H: Microsatellites: simple sequences with complex evolution. Nat Rev Genet 2004, 5(6):435–445.
Evans J, Turnbull JW: Plantation forestry in the tropics. Oxford University Press, Oxford, UK; 2004.
Falahati-Anbaran M, Habashi AA, Esfahany M, Mohammadi SA, Ghareyazie B: Population genetic structure based on SSR markers in alfalfa ( Medigaco sativa L.) from various regions contiguous to the centres of origin of the species. J Genet 2007, 86(1):59–63.
Faria DA, Mamani EM, Pappas MR, Pappas GJ, Grattapaglia DA: Selected set of EST-derived microsatellites, polymorphic and transferable across 6 species of Eucalyptus . J Hered 2010, 101(4):512–520.
Faria DA, Mamani EMC, Pappas GJ, Grattapaglia D: Genotyping systems for Eucalyptus based on tetra-, penta- and hexanucleotide repeat EST microsatellites and their use for individual fingerprinting and assignment tests. Tree Genet Genomes 2011, 7(1):63–77.
Fidalgo M, Barrales RR, Ibeas JI, Jimenez J: Adaptive evolution by mutations in the FLO11 gene. Proc Natl Acad Sci 2006, 103(30):11228–11233.
Field DL, Ayre DJ, Whelan RJ, Young AG: Patterns of hybridization and asymmetrical gene flow in hybrid zones of the rare Eucalyptus aggregate and common E.rubida . Heredity 2010, 106(5):841–853.
Foster SA, McKinnon GE, Steane DA, Potts BM, Vaillancourt RE: Parallel evolution of dwarf ecotypes in the forest tree Eucalyptus globulus . New Phytol 2007, 175(2):370–380.
Freeman JS, Potts BM, Shepherd M, Vaillancourt RE: Parental and consensus linkage maps of Eucalyptus globulus using AFLP and microsatellite markers. Silvae Genet 2006, 55(4–5):202–217.
Freeman JS, Marques CM, Carocha V, Borralho N, Potts BM, Vaillancourt RE: Origins and diversity of the Portuguese Landrace of Eucalyptus globules . Ann Forest Sci 2007, 64(6):639–647.
Freeman JS, O’Reilly-Wapstra M, Vaillancourt RE, Wiggins N, Potts BM: Quantitative trait loci for key defensive compounds affecting herbivory of eucalypts in Australia. New Phytol 2008, 178(4):846–851.
Freeman JS, Potts BM, Vaillancourt RE: Few Mendelian genes underlie the quantitative response of a forest tree, Eucalyptus globulus , to a natural fungal epidemic. Genetics 2008, 178(1):563–571.
Freeman JS, Whittock SP, Potts BM, Vaillancourt RE: QTL influencing growth and wood properties in Eucalyptus globulus . Tree Genet Genomes 2009, 5(4):713–722.
Freeman JS, Potts BM, Downes GM, Pilbeam D, Thavamanikumar S, Vaillancourt RE: Stability of quantitative trait loci for growth and wood properties across multiple pedigrees and environments in Eucalyptus globulus. New Phytol 2013, 198(4):1121–1134.
Garcia M, Villalba P, Acuna C, Oberschelp J, Harrand L, Surenciski M, Poltri SM: A genetic linkage map for a full sib population of Eucalyptus grandis using SSR, DArT, CG-SSR and EST-SSR markers. BMC Proc 2011, 5: 26.
Gauli A, Steane D, Vaillancourt R, Potts B: Molecular genetic diversity and population structure in Eucalyptus pauciflora ssp. pauciflora (Myrtaceae) on the island of Tasmania. Aust J Bot 2014, 62(3):175–188.
Gemayel R, Vinces MD, Legendre M, Verstrepen KJ: Variable tandem repeats accelerate evolution of coding and regulatory sequences. Ann Rev Genet 2010, 44: 445–477.
Gion JM, Carouche A, Deweer S, Bedon F, Pichavant F, Charpentier JP, Bailleres H, Rozenberg P, Carocha V, Ognouabi N, Verhaegen D, Grima-pettenati J, Vigneron P, Plomion C: Comprehensive genetic dissection of wood properties in a widely- grown tropical tree: Eucalyptus . BMC Genomics 2011, 12(1):301.
Glaubitz JC, Emebiri LC, Moran GF: Dinucleotide microsatellites from Eucalyptus sieberi : inheritance, diversity, and improved scoring of single-base differences. Genome 2001, 44(6):1041–1045.
Glaubitz JC, Murrell JC, Moran GF: Effects of native forest regeneration practices on genetic diversity in Eucalyptus consideniana . Theor Appl Genet 2003, 107(3):422–431.
Grattapaglia D: Integrating genomics into Eucalyptus breeding. Genet Mol Res 2004, 3(3):369–379.
Grattapaglia D, Sederoff R: Genetic linkage maps of Eucalyptus grandis and Eucalyptus urophylla using a pseudo-testcross: mapping strategy and RAPD markers. Genetics 1994, 137(4):1121–1137.
Grattapaglia D, Ribeiro VJ, Rezende GDS: Retrospective selection of elite parent trees using paternity testing with microsatellite markers: an alternative short term breeding tactic for Eucalyptus . Theor Appl Genet 2004, 109(1):192–199.
Grattapaglia D, Vaillancourt RE, Shepherd M, Thumma BR, Foley W, Kulheim C, Myburg AA: Progress in Myrtaceae genetics and genomics: Eucalyptus as the pivotal genus. Tree Genet Genomes 2012, 8(3):463–508.
Grattapaglia D, Mamani E, Silva-Junior OB, Faria DA (2014) A novel genome-wide microsatellite resource for species of Eucalyptus with linkage-to-physical correspondence on the reference genome sequence. Mol Ecol Resour. doi: 10.1111/1755–0998.12317 Grattapaglia D, Mamani E, Silva-Junior OB, Faria DA (2014) A novel genome-wide microsatellite resource for species of Eucalyptus with linkage-to-physical correspondence on the reference genome sequence. Mol Ecol Resour. doi: 10.1111/1755-0998.12317
Grosser C, Potts BM, Vaillancourt RE: Microsatellite based paternity analysis in a clonal Eucalyptus nitens seed orchard. Silvae Genet 2008, 59: 57–62.
Gross-German E, Viruel MA: Molecular characterization of avocado germplasm with a new set of SSR and EST-SSR markers: genetic diversity, population structure, and identification of race-specific markers in a group of cultivated genotypes. Tree Genet Genomes 2013, 9(2):539–555.
Hamblin MT, Warburton ML, Buckler ES: Empirical comparison of simple sequence repeats and single nucleotide polymorphisms in assessment of maize diversity and relatedness. PLoS One 2007, 2(12):e1367.
He X, Wang Y, Li F, Weng Q, Li M, Xu LA, Gan S: Development of 198 novels EST- derived microsatellites in Eucalyptus Myrtaceae. Am J Bot 2012, 99(4):134–148.
Healey A, Furtado A, Henry RJ: Structural genomics of eucalypts. In Genetics, genomics and breeding of eucalypts. Edited by: Henry R, Kole C. CRC Press, Boca Raton, FL; 2014:103–120.
Henery ML, Moran GF, Wallis IR, Foley WJ: Identification of quantitative trait loci influencing foliar concentrations of terpenes and formylated phloroglucinol compounds in Eucalyptus nitens . New Phytol 2007, 176(1):82–95.
Hirakawa H, Nakamura Y, Kaneko T, Isobe S, Sakai H, Kato T, Hibino T, Sasamoto S, Watanabe A, Yamada M, Nakayama S, Fujishiro T, Kishida Y, Kohara M, Tabata S, Sato S: Survey of the genetic information carried in the genome of Eucalyptus camaldulensis . Plant Biotech 2011, 28: 471–480.
Hoffman JI, Amos W: Microsatellite genotyping errors: detection approaches, common sources and consequences for paternal exclusion. Mol Ecol 2005, 14(2):599–612.
Holman JE, Hughes JM, Fensham RJ: A morphological cline in Eucalyptus : a genetic perspective. Mol Ecol 2003, 12(11):3013–3025.
Hudson CJ, Freeman JS, Kullan AR, Petroli CD, Sansaloni CP, Kilian A, Vaillancourt RE: A reference linkage map for Eucalyptus . BMC Genomics 2012, 13(1):240.
Jones ME, Stokoe RL, Cross MJ, Scott LJ, Maguire TL, Shepherd M: Isolation of microsatellite loci from spotted gum Corymbia variegate and cross-species amplification in Corymbia and Eucalyptus . Mol Ecol Notes 2001, 1(4):276–278.
Jones RC, Steane DA, Potts BM, Vaillancourt RE: Microsatellite and morphological analysis of Eucalyptus globulus populations. Can J Forest Res 2002, 32(1):59–66.
Jones TH, Steane DA, Jones RC, Pilbeam D, Vaillancourt RE, Potts BM: Effects of domestication on genetic diversity in Eucalyptus globulus . For Ecol Manag 2006, 234(1):78–84.
Jones TH, Vaillancourt RE, Potts BM: Detection and visualization of spatial genetic structure in continuous Eucalyptus globulus forest. Mol Ecol 2007, 16(4):697–707.
Jones ME, Shepherd M, Henry R, Delves A: Pollenflow in Eucalyptus grandis determined by paternity analysis using microsatellite markers. Tree Genet Genomes 2008, 4(1):37–47.
Jones RC, Steane DA, Lavery M, Vaillancourt RE, Potts BM: Multiple evolutionary processes drive the patterns of genetic differentiation in a forest tree species complex. Ecol Evol 2013, 3(1):1–17.
Joy N, Soniya EV: Identification of an miRNA candidate reflects the possible significance of transcribed microsatellites in the hairpin precursors of black pepper. Funct Integr Genom 2012, 12(2):387–395.
Joy N, Asha S, Mallika Vand Soniya EV: De novo transcriptome sequencing reveals a considerable bias in the incidence of simple sequence repeats towards the downstream of `pre-miRNAs’ of black pepper. PLoS One 2013, 8(3):e56694.
Kalia RK, Rai MK, Kalia S, Singh R, Dhawan AK: Microsatellite markers: an overview of the recent progress in plants. Euphytica 2011, 177(3):309–334.
Khadivi-Khub A, Zamani Z, Fattahi R, Wünsch A: Genetic variation in wild Prunus L. subgen. Cerasus germplasm from Iran characterized by nuclear and chloroplast SSR markers. Trees 2014, 28(2):471–485.
Kirst M, Myburg AA, De Leon JP, Kirst ME, Scott J, Sederoff R: Coordinated genetic regulation of growth and lignin revealed by quantitative trait locus analysis of cDNA microarray data in an interspecific backcross of Eucalyptus . Plant Physiol 2004, 135(4):2368–2378.
Kirst M, Basten CJ, Myburg AA, Zeng ZB, Sederoff RR: Genetic architecture of transcript-level variation in differentiating xylem of a Eucalyptus hybrid. Genetics 2005, 169(4):2295–2303.
Kirst M, Cordeiro CM, Rezende G, Grattapaglia D: Power of microsatellite markers for fingerprinting and parentage analysis in Eucalyptus grandis breeding populations. J Heredity 2005, 96(2):161–166.
Kudapa H, Azam S, Sharpe AG, Taran B, Li R, Deonovic B, Varshney RK: Comprehensive transcriptome assembly of chickpea Cicer arietinum L . Using Sanger and next generation sequencing platforms: development and applications. PLoS One 2014, 9(1):e86039.
Kullan ARK, Van Dyk MM, Jones N, Kanzler A, Bayley A, Myburg AA: High - density genetic linkage maps with over 2,400 sequence-anchored DArT markers for genetic dissection in an F2 pseudo-backcross of Eucalyptus grandis × E. urophylla . Tree Genet Genomes 2011, 8(1):163–175.
Kullan ARK, Van Dyk MM, Hefer CA, Jones N, Kanzler A, Myburg AA: Genetic dissection of growth, wood basicdensity and gene expression in interspecific backcrosses of Eucalyptus grandis and E. urophylla . BMC Genet 2012, 13(1):60.
Luo J, Arnold R, Ren S, Jiang Y, Lu W, Peng Y, Xie Y: Veneer grades, recoveries and values from 5-year old eucalypt clones. Ann For Sci 2013, 70(4):417–428.
Mamaghani MS, Shojaei TR, Matinizadeh M, Forootan M: Microsatellite loci and peroxidase alleles correlation in somaclonal variation of Eucalyptus microtheca F. Muell Afr J Biotechnol 2010, 9(29):4521–4528.
Mamani EM, Bueno NW, Faria DA, Guimaraes LM, Lau D, Alfenas AC, Grattapaglia D: Positioning of the major locus for Puccinia psidii rust resistance Ppr1 on the Eucalyptus reference map and its validation across unrelated pedigrees. Tree Genet Genomes 2010, 6(6):953–962.
Marques CM, Brondani RPV, Grattapaglia D, Sederoff R: Conservation and synteny of SSR loci and QTLs for vegetative propagation in four Eucalyptus species. Theor Appl Genet 2002, 105(2–3):474–478.
Missiaggia A, Grattapaglia D: Plant microsatellite genotyping with 4-color fluorescent detection using multiple-tailed primers. Genet Mol Res 2006, 5(1):72–78.
Missiaggia AA, Piacezzi AL, Grattapaglia D: Genetic mapping of Eef1, a major effect QTL for early flowering in Eucalyptus grandis . Tree Genet Genomes 2005, 1(2):79–84.
Mizrachi E, Hefer C, Ranik M, Joubert F, Myburg AA: De novo assembled expressed gene catalog of a fast-growing Eucalyptus plantation tree produced by Illumina mRNA- seq. BMC Genomics 2010, 11(1):681.
Mondal TK, Ganie SA: Identification and characterization of salt responsive miRNA- SR markers in rice ( Oryza sativa ). Gene 2014, 535(2):204–209.
Myburg AA, Potts BM, Marques CM, Kirst M, Gion JM, Grattapaglia D, Grima-Pettenati J: Eucalyptus. In Genome mapping and molecular breeding in plants. Edited by: Kole C. Springer, NY; 2007:115–160.
Myburg AA, Grattapaglia D, Tuskan GA, Hellsten U, Hayes RD, Grimwood J, Jenkins J, Lindquist E, Tice H, Bauer D, Goodstein DM, Dubchak I, Poliakov A, Mizrachi E, Kullan ARK, Hussey SG, Pinard D, van der Merwe K, Singh P, van Jaarsveld I, Silva-Junior OB, Togawa RC, Pappas MR, Faria DA, Sansaloni CP, Petroli CD, Yang X, Priya R, Tschaplinski TJ, Chu-Yu Y, et al.: The genome of Eucalyptus grandis . Nature 2014, 510(7505):356–362.
Nagabhushana K, Hendre PS, Sharma N, Rathinavelu R: Novel design and deployment of orthologous genic SSR markers in Eucalyptus camaldulensis Dehnh. BMC Proc 2011, 5(Suppl 7):51.
Neves LG, Mamani EM, Alfenas AC, Kirst M, Grattapaglia D: A high-density transcript linkage map with 1,845 expressed genes positioned by microarray-based Single Feature Polymorphisms SFP in Eucalyptus . BMC Genomics 2011, 12(1):189.
Nevill PG, Reed A, Bossinger G, Vaillancourt RE, Larcombe M, Ades PK: Cross-species amplification of Eucalyptus microsatellite loci. Mol Ecol Res 2008, 8(6):1277–1280.
Nevill PG, Bossinger G, Ades PK: Phylogeography of the world’s tallest angiosperm, Eucalyptus regnans : evidence for multiple isolated Quaternary refugia. J Biogeogr 2010, 37(1):179–192.
Nevill PG, Bradbury D, Jorgensen T, Krauss S, Samaraweera S, Gardner MG (2013) Microsatellite primers identified by 454 sequencing in the floodplain tree species Eucalyptus victrix (Myrtaceae). Appl Plant Sci 1(5):, [http://dx.doi.org/10.3732/apps.1200402] Nevill PG, Bradbury D, Jorgensen T, Krauss S, Samaraweera S, Gardner MG (2013) Microsatellite primers identified by 454 sequencing in the floodplain tree species Eucalyptus victrix (Myrtaceae). Appl Plant Sci 1(5):,
Nevill PG, Bradbury D, Williams A, Tomlinson S, Krauss SL: Genetic and palaeo- climatic evidence for widespread persistence of the coastal tree species Eucalyptus gomphocephala (Myrtaceae) during the last glacial maximum. Ann Bot 2014, 113(1):55–67.
Ochieng J, Steane DA, Ladiges PY, Baverstock PR, Henry RJ, Shepherd M: Microsatellites retain phylogenetic signals across genera in eucalypts (Myrtaceae). Genet Mol Biol 2007, 30(4):1125–1134.
Ottewell KM, Donnellan SC, Moran GF, Paton DC: Multiplexed microsatellite markers for the genetic analysis of Eucalyptus leucoxylon Myrtaceae and their utility for ecological and breeding studies in other Eucalyptus species. J Heredity 2005, 96(4):445–451.
Paiva JA, Prat E, Vautrin S, Santos MD, Clemente SH, Brommonschenkel S, Grima- Pettenati J: Advancing Eucalyptus genomics: identification and sequencing of lignin biosynthesis genes from deep-coverage BAC libraries. BMC Genomics 2011, 12(1):137.
Patterson B, Vaillancourt RE, Pilbeam DJ, Potts BM: Factors affecting variation in outcrossing rate in Eucalyptus globulus . Aust J Bot 2004, 52(6):773–780.
Payn KG, Dvorak WS, Janse BJH, Myburg AA: Microsatellite diversity and genetic structure of the commercially important tropical tree species Eucalyptus urophylla , endemic to seven islands in eastern Indonesia. Tree Genet Genomes 2008, 4(3):519–530.
Petroli CD, Sansaloni CP, Carling J, Steane DA, Vaillancourt RE, Myburg AA, Grattapaglia D: Genomic characterization of DArT markers based on high- density linkage analysis and physical mapping to the Eucalyptus genome. PLoS One 2012, 7(9):e44684.
Poltri SM, Zelener N, Traverso JR, Gelid P, Hopp HE: Selection of a seed orchard of Eucalyptus dunnii based on genetic diversity criteria calculated using molecular markers. Tree Physiol 2003, 23(9):625–632.
Porth I, El-Kassaby YA: Assessment of the genetic diversity in forest tree populations using molecular markers. Diversity 2014, 6(2):283–295.
Rabello E, Souza NDA, Saito D, Tsai MS: In silico characterisation of microsatellites in Eucalyptus spp.: abundance, length variation and transposon associations. Genet Molar Biol 2005, 28(3):582–588.
Ranade SS, Lin YC, Zuccolo A, Van de Peer Y, García-Gil MR: Comparative in silico analysis of EST-SSRs in angiosperm and gymnosperm tree genera. BMC Plant Biol 2014, 14(1):220.
Rao HX, Patterson B, Potts B, Vaillancourt R: A microsatellite study on outcrossing rates and contamination in an Eucalyptus globulus breeding arboretum. J For Res 2008, 19(2):136–140.
Rathbone DA, McKinnon GE, Potts BM, Steane DA, Vaillancourt RE: Microsatellite and cpDNA variation in island and mainland populations of a regionally rare eucalypt, Eucalyptus perriniana Myrtaceae. Aust J Bot 2007, 55(5):513–520.
Rengel D, Clemente HS, Servant F, Ladouce N, Paux E, Wincker P, Couloux A, Sivadon P, Grima-Pettenati J: A new Eucalyptus genomic resource dedicated to wood formation: a comprehensive survey. BMC Plant Biol 2009, 9(1):36.
Ribeiro MM, Sanchez L, Ribeiro C, Cunha F, Araujo J, Borralho NM, Marques C: A case study of Eucalyptus globulus fingerprinting for breeding. A Forest Science 2011, 68(4):701–714.
Rosado TB, Tomaz RS, Ribeiro Junior MF, Rosado AM, Guimaraes LMDS, Araujo EFD, Cruz CD: Detection of QTL associated with rust resistance using IBD-based methodologies in exogamic Eucalyptus spp. populations. Crop Breed Appl Biotechnol 2010, 10(4):321–328.
Sampson JF, Byrne M: Outcrossing between an agroforestry plantation and remnant native populations of Eucalyptus loxophleba . Mol Ecol 2008, 17(11):2769–2781.
Sexton TR, Henry RJ, McManus LJ, Bowen S, Shepherd M: Capture of assay template by multiplex PCR of long amplicons for genotyping SNPs and InDels with MALDI-TOF mass spectrometry. Mol Breed 2010, 25(3):471–480.
Shepherd M, Raymond CA: Species delineation and gene flow in the Blackbutts genus Eucalyptus subgenus Eucalyptus section Pseudophloius. Conserv Genet 2010, 11(5):1965–1978.
Shepherd M, Kasem S, Lee D, Henry R: Construction of microsatellite linkage maps for Corymbia . Silvae Genet 2006, 55(4–5):228–237.
Shepherd M, Kasem S, Lee DJ, Henry R: Mapping species differences for adventitious rooting in a Corymbia torelliana × Corymbia citriodora subspecies variegata hybrid. Tree Genet Genomes 2008, 4(4):715–725.
Shepherd M, Sexton TR, Thomas D, Henson M, Henry RJ: Geographical and historical determinants of microsatellite variation in Eucalyptus pilularis . Can J Forest Res 2010, 40(6):1051–1063.
Shi L, Lv X, Weng J, Zhu H, Liu C, Hao Z, Zhang S: Genetic characterization and linkage disequilibrium mapping of resistance to gray leaf spot in maize ( Zea mays L). The Crop Journal 2014, 2(2):132–143.
Sitther V, Zhang D, Harris DL, Yadav AK, Zee FT, Meinhardt LW, Dhekney SA: Genetic characterization of guava ( Psidium guajava L.) germplasm in the United States using microsatellite markers. Genetic Resour Crop Evol 2014, 61(4):829–839.
Smith S, Hughes J, Wardell-Johnson G: High population differentiation and extensive clonality in a rare mallee eucalypt: Eucalyptus curtisii - conservation genetics of a rare mallee eucalypt. Conserv Genet 2003, 4: 289–300.
Steane D, Vaillancourt R, Russell J, Powell W, Marshall D, Potts B: Development and characterisation of microsatellite loci in Eucalyptus globulus Myrtaceae. Silvae Genet 2001, 50(2):89–92.
Steane DA, Jones RC, Vaillancourt RE: A set of chloroplast microsatellite primers for Eucalyptus Myrtaceae. Mol Ecol Notes 2005, 5(3):538–541.
Steane DA, Conod N, Jones RC, Vaillancourt RE, Potts BM: A comparative analysis of population structure of a forest tree, Eucalyptus globulus Myrtaceae, using microsatellite markers and quantitative traits. Tree Genet Genomes 2006, 2(1):30–38.
Stokoe RL, Shepherd M, Lee D, Nikles DG, Henry RJ: Natural interspecific hybridization between Eucalyptus acmenoides Schauer and E. cloeziana F. Muell Myrtaceae. Ann Bot 2001, 88(4):563–570.
Subashini V, Shanmugapriya A, Yasodha R: Hybrid purity assessment in Eucalyptus F1hybrids using microsatellite markers. 3 Biotech 2013, 4(4):367–373.
Teixeira J, Missiaggia A, Dias D, Scarpinati E, Viana J, Paula N, Paula R, Bonine C: QTL analyses of drought tolerance in Eucalyptus under two contrasting water regimes. BMC Proc 2011, 5: 40.
Terzoli S, Abbruzzese G, Beritognolo I, Sabatti M, Valentini R, Kuzminsky E: Genetic characterization of a Tamarix spp. germplasm collection in Italy. Botany 2014, 92(5):360–369.
Thamarus K, Groom K, Murrell J, Byrne M, Moran G: A genetic linkage map for Eucalyptus globulus with candidate loci for wood, fibre and floral traits. Theor Appl Genet 2002, 104(2–3):379–387.
Thumma BR, Southerton SG, Bell JC, Owen JV, Henery ML, Moran GF: Quantitative trait locus QTL analysis of wood quality traits in Eucalyptus nitens . Tree Genet Genomes 2010, 6(2):305–317.
Tripiana V, Bourgeois M, Verhaegen D, Vigneron P, Bouvet JM: Combining microsatellites, growth, and adaptive traits for managing in situ genetic resources of Eucalyptus urophylla . Can J For Res 2007, 37(4):773–785.
Trujillo I, Ojeda MA, Urdiroz NM, Potter D, Barranco D, Rallo L, Diez CM: Identification of the Worldwide Olive Germplasm Bank of Córdoba (Spain) using SSR and morphological markers. Tree Genet Genomes 2014, 10(1):141–155.
Van Der Nest MA, Steenkamp ET, Wingfield BD, Wingfield MJ: Development of simple sequence repeat SSR markers in Eucalyptus from amplified inter-simple sequence repeats ISSR. Plant Breed 2000, 119(5):433–436.
van Zonneveld M, Scheldeman X, Escribano P, Viruel MA, Van Damme P, Garcia W, Tapia C, Romero J, Sigueñas M, Hormaza JH: Mapping genetic diversity of cherimoya ( Annona cherimola Mill.): Application of spatial analysis for conservation and use of plant genetic resources. Plos One 2012, 7: e29845.
van Zonneveld M, Dawson I, Thomas E, Scheldeman X, van Etten J, Loo J, Hormaza JI: Application of molecular markers in spatial analysis to optimize in situ conservation of plant genetic resources. Genomics of plant genetic resources. Springer, Netherlands; 2014.
Varshney RK, Graner A, Sorrells ME: Genic microsatellite markers in plants: features and applications. Trends in Biotech 2005, 23: 48–55.
Wang ML, Barkley NA, Jenkins TM: Microsatellite markers in plants and insects. Part I applications of biotechnology. Genes Genomes Genomics 2009, 3(1):54–67.
Wheeler R, Nevill PG, Renton M, Krauss SL: Interspecific hybridisation in tuart Eucalyptus gomphocephala , Myrtaceae: a conservation management issue? Aust J Bot 2013, 61(6):455–464.
Yang X, Xu Y, Shah T, Li H, Yan J: Comparison of SSRs and SNPs in assessment of genetic relatedness in maize. Genetica 2011, 139(8):1045–1054.
Yasodha R, Sumathi R, Chezhian P, Kavitha S, Ghosh M: Eucalyptus microsatellites mined in silico: survey and evaluation. J Genet 2008, 87(1):21–25.
Yeoh SH, Bell JC, Foley WJ, Wallis IR, Moran GF: Estimating population boundaries using regional and local-scale spatial genetic structure: an example in Eucalyptus globules . Tree Genet Genomes 2012, 8(4):695–708.
Zalapa JE, Cuevas H, Zhu H, Steffan S, Senalik D, Zeldin E, Simon P: Using next- generation sequencing approaches to isolate simple sequence repeat SSR loci in the plant sciences. Am J Bot 2012, 99(2):193–208.
Zelener N, Poltri SN, Bartoloni N, Lopez CR, Hopp HE: Selection strategy for a seedling seed orchard design based on trait selection index and genomic analysis by molecular markers: a case study for Eucalyptus dunnii . Tree Physiol 2005, 25(11):1457–1467.
Zhou C, Li F, Weng Q, Yu X, Li M, Gan S: Comparison between direct sequencing and pool-cloning-based sequencing of PCR products in EST-SSR marker development in Eucalyptus . Mol Plant Breed 2010, 8: e1. doi:10.5376/mpb.cn.2010.08.0001 doi:10.5376/mpb.cn.2010.08.0001
Zhou C, He X, Li F, Weng Q, Yu X, Wang Y, Gan S: Development of 240 novel EST- SSRs in Eucalyptus L’Herit.. Mol Breed 2014, 33(1):221–225.
Acknowledgements
The first author acknowledges the Junior Research Fellowship from the Department of Biotechnology, Government of India.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
MS surveyed the literature and drafted the manuscript. RY provided guidence for the review, modified and prepared the final version of the manuscript. Both the authors read and approved the final manuscript.
Electronic supplementary material
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0), which permits use, duplication, adaptation, distribution, and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Sumathi, M., Yasodha, R. Microsatellite resources of Eucalyptus: current status and future perspectives. Bot Stud 55, 73 (2014). https://doi.org/10.1186/s40529-014-0073-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40529-014-0073-3