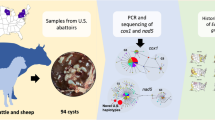
Abstract
Background
Canids are definitive hosts of Echinococcus multilocularis and Echinococcus granulosus. This study aimed to survey these two Echinococcus species in canids of North-Khorasan Province, northeastern Iran, using morphological criteria and genetic characterization of mitochondrial DNA.
Methods
The carcasses of 106 canids, namely 61 jackals (Canis aureus), 23 foxes (Vulpes vulpes), 19 dogs (Canis familiaris) and three wolves (Canis lupus) were collected from the study area in 2013–2014 and examined for Echinococcus species. Morphological features were assessed by microscopy of adult worms. For molecular characterization, DNA was extracted, mostly from the adult worms but also from eggs. DNA fragments of the cytochrome c oxidase subunit 1 (cox1) and NADH dehydrogenase subunit 1 (nad1) mitochondrial genes were amplified and sequenced. Sequences were aligned and compared with reference sequences. Intraspecific and interspecific diversity were calculated and phylogenetic analysis was performed.
Results
Overall, 9.4% of the canids (eight jackals and two foxes) were found infected with E. multilocularis by molecular methods, of which seven cases were also confirmed using morphological description of the adult worms. Echinococcus granulosus was found in 6.6% of the canines (four dogs, two jackals and one wolf) as determined by both molecular methods and adult cestode morphology. All E. granulosus isolates were identified as the G1 genotype. Comparative sequence analysis indicated 0–0.7% and 0% intraspecific divergence within E. granulosus isolates and 0% and 0–0.2% within E. multilocularis isolates for cox1 and nad1, respectively.
Conclusions
This study revealed the presence of E. multilocularis and E. granulosus in canids of North-Khorasan Province of Iran. Jackals were found infected with both E. multilocularis and E. granulosus, but infection with the former species was higher.

Similar content being viewed by others
Background
Echinococcosis, caused by the larval stage of tapeworms of the genus Echinococcus, is one of the most important zoonotic diseases worldwide [1]. Echinococcus granulosus (sensu lato) and Echinococcus multilocularis are the most prevalent species infecting humans, resulting in cystic echinococcosis (CE) and alveolar echinococcosis (AE), respectively. Echinococcus granulosus (s.l.) is known to be endemic in all continents, while E. multilocularis has a more restricted distribution, generally regarded a parasite limited to the northern hemisphere [2]. Both AE and CE are considered neglected zoonoses, with a global distribution and higher prevalence for CE, but a higher pathogenicity and mortality for AE, especially in Asia [3]. Herbivores are intermediate hosts for E. granulosus (s.l.), and canids, including dogs, wolves, foxes and jackals, act as definitive hosts, harboring the adult worms in the villi of the small intestine [4]. The life-cycle of E. multilocularis involves several carnivores such as foxes, coyotes, dogs and cats as definitive hosts, and rodents as intermediate hosts. Humans can be an accidental dead-end intermediate host for both species via close contact with the definitive host or by indirect ingestion of eggs through contaminated water and uncooked food [5].
Without data on genetic variation within and between populations of Echinococcus, no decisions can be made about breeding systems, extent of gene flow, species delineation or modes of speciation [6]. During the past decades, molecular studies, mainly based on mitochondrial genes, have described several genotypes or species within E. granulosus (s.l.), revealing a species complex as follows: E. granulosus (sensu stricto) (genotypes G1–G3), E. equinus (G4), E. ortleppi (G5), E. canadensis (G6–G10) and E. felidis (‘lion strain’); the existence of the human-specific genotype G9 is controversial [1, 7]. Recently, Kinkar et al. [8] showed that G1 and G3 are two distinct mitochondrial genotypes and can be considered as a single species of E. granulosus (s.s.), whereas G2 is not a separate genotype but belongs to G3. They suggested eliminating G2 from the list of E. granulosus genotypes. Laurimae et al. [9] confirmed that based on six nuclear loci, G6/G7 and G8/G10 genotypes can be considered as two distinct species. Additionally, Thompson [10] proposed to consider camel and pig strains of E. granulosus as a single species (E. intermedius) as originally suggested by Lopez-Neyra & Soler Planas in 1943 [11]. Unlike E. granulosus (s.l.), only minor variations have been detected in the cytochrome c oxidase subunit 1 (cox1) and NADH dehydrogenase subunit 1 (nad1) mitochondrial DNA sequences of E. multilocularis isolates, resulting in the recognition of two groups, namely M1 and M2. M1 originates in China and North America and M2 in Europe [12, 13].
To date, molecular studies on E. granulosus carried out in Iran have reported several genotypes in domestic livestock (genotypes G1–G7) [14,15,16,17,18] as well as in humans (G1–G3 and G6) from different endemic foci of Iran [19,20,21,22,23]. Additionally, the genotypes G1 [24,25,26,27], G2 [24], G3 [24,25,26] and G6 [25] have been identified in dogs in Iran.
So far, several documented human cases of alveolar echinococcosis have been reported in different parts of Iran [28,29,30]. Canine infection with adult worms of E. multilocularis has been previously reported in red foxes [31, 32] and jackals [32] from northwestern Iran based on morphological criteria. Furthermore, E. multilocularis has been reported in carnivores from the Razavi Khorasan Province, northeastern Iran, using analysis of nad1 mitochondrial DNA [33].
In Iran, domestic dogs, but also wild canids including foxes, jackals and wolves, are known as important reservoirs of echinococcosis [34]. However, information on the role of such animals in the spread of this disease is available for only some provinces. The North-Khorasan Province, located in northeastern Iran, is bordering with Turkmenistan where E. multilocularis is believed to be endemic [35]. There are reports of human CE in North-Khorasan Province [36,37,38]. There are also reports of E. multilocularis in carnivores in Chenaran City in the adjacent province, Razavi Khorasan [33]. Additionally, a variety of rodent species, including the family Cricetidae, members of which act as the main natural intermediate hosts of AE in other endemic parts of the world [39], have also been observed in northeastern Iran [40]. Nevertheless, at present there are no data available on Echinococcus spp. in the canids of this province. Therefore, the purpose of this study was to identify species and genotypes of Echinococcus spp. in canids of North-Khorasan Province using morphological criteria and sequencing partial cox1 and nad1 genes.
Methods
Study area
North-Khorasan Province is located between 36°37′–38°17′ N and 55°53′–58°20′ E, comprising an area of 28,434 km2 in northeastern Iran and sharing a border with Turkmenistan (Fig. 1). The capital is Bojnord. The province has a temperate climate with cold winters. The variety of different climates in this province is because of the vast mountains and forests. This region is geographically divided into two parts: mountain areas and lowland areas.
Sampling
Study samples comprised carcasses of 106 canids [61 golden jackals (Canis aureus); 23 red foxes (Vulpes vulpes); 19 dogs (Canis familiaris); and three gray wolves (Canis lupus)] provided by the Vector-Borne Diseases Research Center (VDRC) in Bojnord. The carcasses were collected from 2013 to 2014 and reflected mostly road-kill accidents in different locations of the province. Other carcasses were part of the collection from other research projects in the same area on visceral leishmaniasis in wild canines, in collaboration with the VDRC [41].
In the VDRC, a veterinary practitioner recorded the characteristics of each animal including age group and sex in the relevant registration form with a specific identification code. The approximate age of the animals was determined according to the shape and color of teeth and dental formula to three groups (cub, under 2 years; young, 2–5 years; adult, > 5 years).
Under safety precautions, every carcass was dissected, and the entire length of the small intestine tied off, removed and stored in plastic container in a mixture of 70% ethanol (ethanol and water, 70:30, v/v). Fecal samples were also prepared from the rectum of the canids and kept in separate plastic containers with 70% ethanol. All samples were transported to the School of Public Health, Tehran University of Medical Sciences, Iran, and stored at − 20 °C for at least one month prior to examination.
Parasitological examinations
For examination of the small intestines, the intestinal scraping technique (IST) was applied under safety precautions as described by Deplazes & Eckert [42]. In brief, each small intestine was opened along its full length in a metal tray, and after removal of coarse materials and large helminths, deep mucosal scrapings were made from the proximal, middle and posterior third of the small intestine using microscopic slides. The adhering materials were transferred to plastic Petri dishes and examined for adult Echinococcus tapeworms under a stereomicroscope. Additionally, small intestinal content and the remaining mucosa were washed with water through a sieve and the collected precipitations were examined in the same way for detection of Echinococcus. After detection, adult worms were removed, and for each intestine, ten worms were prepared for morphological diagnosis, after clearing in lactophenol and temporary staining using FAAL stain (formalin, alcohol, azocarmine and lactophenol) [43].
To detect taeniid eggs in feces, all fecal samples were examined by the formalin-ether sedimentation technique [44]. If an animal was negative for adult Taeniidae (Echinococcus spp. or Taenia spp.) by IST, but stool-positive for taeniid eggs, the correspondent fecal sample was later processed for molecular analysis.
The morphological characteristics of Echinococcus adult worms were studied using a calibrated microscope with consideration of specific criteria including body length, appearance of rostellar hooks (shape, total length and blade length of large and small hooks), and features of the strobila such as the position of genital pore in proglottids and the shape of the uterus. Species identification was performed according to published guidelines [45, 46]. One adult worm per intestine was kept in 70% ethanol at − 20 °C for further molecular analysis.
Molecular analysis
DNA extraction
For each canid from which adult Echinococcus spp. were recovered by intestinal examination, genomic DNA was extracted from a single morphologically identified worm. Additionally, a DNA extraction protocol was performed on those fecal samples in which taeniid eggs were detected but where no adult taeniid worms could be found in the intestine.
After removal of the ethanol from adult Echinococcus worms and from fecal samples, these were washed twice with sterile distilled water, and total genomic DNA was extracted using a Bioneer tissue DNA extraction kit and stool DNA extraction kit (Bioneer, Daejeon, South Korea), respectively, according to manufacturer’s instructions. All extracted DNA was stored at − 20 °C until PCR amplification.
Polymerase chain reaction (PCR)
Genomic DNA samples were analyzed using amplification of mitochondrial DNA within the cytochrome c oxidase subunit 1 (cox1) and NADH dehydrogenase subunit 1 (nad1) genes, separately. All PCR reactions were carried out in a final volume of 25 μl, consisting of 12.5 μl of master mix (2× Master Mix RED; Ampliqon, Odense, Denmark; 1.25 U Taq DNA polymerase, 0.5 μM of dNTPs and 2 mM MgCl2), 20 pmol of each primer and 2 μl of DNA template. The forward and reverse primers used in the PCRs were as follows: JB3 (5′-TTT TTT GGG CAT CCT GAG GTT TAT-3′) and JB4.5 (5′-TAA AGA AAG AAC ATA ATG AAA ATG-3′) for the cox1 gene [12] and JB11 (5′-AGA TTC GTA AGG GGC CTA ATA-3′) and JB12 (5′-ACC ACT AAC TAA TTC ACT TTC-3′) for the nad1 gene [13]. The PCR program began with one cycle at 94 °C for 5 min (primary denaturation), followed by 35 cycles at 94 °C for 30 s (denaturation), 50 °C (nad1) or 55 °C (cox1) for 45 s (annealing) and 72 °C for 35 s (extension), followed by a final extension step at 72 °C for 10 min.
PCR products were electrophoresed on a 1.5% TBE (Tris 0.09 M, Borate 0.09 M, EDTA 0.02 M) agarose gel stained with Fluoro Dye Fluorescent DNA Loading Dye for loading and detecting DNA markers (SMOBiO DM3100; Bioshimigene, Tehran, Iran). Electrophoresis was carried out at 90 V for 45 min. PCR products were visualized using a UV transilluminator (Uvitec, Cambridge, UK) and digitally photographed. All PCR products of both cox1 and nad1 genes were purified with an AccuPrep Gel purification kit (Bioneer, Daejeon, South Korea) according to the manufacturer’s instructions.
Sequencing and phylogenetic analysis
Purified PCR products were first sequenced unidirectionally using the forward primers indicated above. After analysis of the results, if necessary, the process would be repeated to obtain desirable sequences. Sequences were edited and analyzed by Chromas software v.2.01 (Technelysium Pty Ltd., Brisbane, Queensland, Australia). Nucleotide sequences were compared with reference sequences in GenBank using the BLAST algorithm (http://www.ncbi.nlm.nih.gov/). In addition, sequences were trimmed, aligned using the software BioEdit v.7.0.9 [47], and compared with reference genotypes (G1–G10) of E. granulosus (s.l.), and species of Echinococcus from previous publications. Different nucleotide sequences of both cox1 and nad1 genes from this study were submitted to the GenBank database.
Phylogenetic analysis was performed of representative concatenated cox1 and nad1 DNA sequence data from the present study along with reference sequences of all known E. granulosus genotypes (G1–G10) and Echinococcus species, using Taenia saginata as the outgroup. The character-based Bayesian inference (BI) method was employed for the phylogenetic analyses using the software MrBayes v.3.1.2 [48]. Posterior probabilities (pp) were obtained for 1,000,000 generations (ngen: 1,000,000; ‛burn inʼ = 10,000). The program TreeviewX v.0.5.0 [49] was used to show the consensus tree.
Results
Parasitological and molecular findings
A total of 106 canid intestines were examined for infection with Echinococcus species by morphological and molecular methods. In Table 1, study animals have been listed according to sex and age group.
Overall, 17 of 106 canids (16%) were infected with Echinococcus species. Morphological species identification was carried out based on the characteristics of scolex and strobila. Figures 2, 3, 4, 5, 6 represent comparative morphological characteristics of E. multilocularis and E. granulosus as demonstrated by light microscopy. In the strobila, the total length and proglottid length of mature worms and position of the genital pore (Fig. 2), as well as the shape of the uterus in gravid proglottids (Fig. 3), were discriminative. However, in immature worms, there was an overlap in the size of strobila and proglottids. In the scolex, the dimensions of the suckers (Fig. 4) and of the large and small hooks as well as their shape (Figs. 5, 6) allowed the differentiation between E. multilocularis and E. granulosus. However, the number of hooks could not be counted accurately in all samples due to partial hook detachment during technical processing. Based on the morphology of adult worms, 14 animals were found infected with Echinococcus species: E. multilocularis (n = 7); and E. granulosus (n = 7) (Table 2). No co-infection with these species was found by morphological analysis of ten adult samples of Echinococcus from each infected animal.
For each canid from which adult Echinococcus was retrieved (n = 14), one morphologically described worm was processed for molecular analysis. Moreover, four fecal samples containing taeniid eggs for which corresponding intestinal examination failed to reveal any adult taeniids were also subjected to molecular investigation. Among these four samples, three cases were identified as E. multilocularis (Table 2) and the remaining one as Taenia hydatigena. Analysis of cox1 and nad1 genes of the adult Echinococcus spp. revealed the presence of both E. granulosus and E. multilocularis in the canids from the study area. In all mentioned Echinococcus isolates, fragments of approximately 450 and 500 bp were successfully amplified for cox1 and nad1 genes, respectively. Molecular identification based on both genes was in accordance with morphological identification of the adult worms.
Among the 106 animals examined, E. multilocularis and E. granulosus were identified in ten and seven canids, respectively. All isolates of E. granulosus were identified as the G1 genotype. Table 2 summarizes molecular and morphological results for all Echinococcus isolates detected from different canines, according to sex and age group of the animals. The only animal species hosting both E. multilocularis and E. granulosus was the jackal; however, infectivity with E. multilocularis was higher (13.1 vs 3.3%). Among other canines, 8.7% of the foxes were found infected with E. multilocularis, while 21% of the dogs had E. granulosus. Finally, one out of three wolves was infected with E. granulosus. All infected animals were male animals (Table 2). However, it should be noticed that the sex balance of the study sample was skewed (83 males vs 23 females).
Among the E. granulosus isolates, two different cox1 sequences (scox1-1 and scox1-2) were found, while all nad1 sequences were identical (snad1-1). Among the E. multilocularis isolates, two different nad1 sequences (snad1-2 and snad1-3) were detected (intraspecific diversity of 0–0.2%), while all cox1 sequences were identical (scox1-3). Comparative sequence analysis showed 0–0.7% and 0% intraspecific genetic divergence within E. granulosus isolates and 0% and 0–0.2% within E. multilocularis isolates for cox1 and nad1, respectively. All 17 partial DNA sequences of cox1 and nad1 genes obtained in this study were deposited in the GenBank database under the accession numbers shown in Table 3.
Phylogenetic analysis
A rooted phylogenetic tree using T. saginata as the outgroup was constructed based on concatenated data of 600 nucleotides (nt) including cox1 (282 nt) and nad1 (318 nt) using Bayesian inference. Overall, two haplotypes of E. granulosus (s.s.), referred to as E.gKh63 and E.gKh75, and two haplotypes of E. multilocularis, referred to as E.mKh2 and E.mKh4, were observed in the present study. Representative haplotypes of E. granulosus clustered with a strong support (pp = 1.00) with G1–G3 genotypes, and representative haplotypes of E. multilocularis clustered with a strong support (pp = 1.00) with the reference sequences of E. multilocularis (Fig. 7).
Phylogenetic tree of Echinococcus granulosus and Echinococcus multilocularis isolates from North-Khorasan Province, northeastern Iran (indicated in bold) and reference sequences for E. granulosus (sensu lato) and other species of Echinococcus chosen from previous studies. The relationships were estimated based on phylogenetic analysis of concatenated cox1+nad1 sequence data (600 nucleotides in total) using Bayesian inference; the sources and accession numbers of the sequences are listed in Table 3. The scale-bar indicates the number of substitutions per site. Nodal support is given as a posterior probability (pp) value. Taenia saginata was used as the outgroup
Discussion
Echinococcosis is one of the main neglected zoonotic diseases [50]. Echinococcus granulosus (s.l.) is known to exist in all continents, while E. multilocularis is displaying endemic districts in central Europe, northern and central Eurasia and particular parts in North America [2]. With regard to the Middle East, Iran is known as one of the endemic countries for both E. granulosus and E. multilocularis [34]. Echinococcus granulosus is highly prevalent throughout the country [34], and E. multilocularis is endemic in part of the territory in the north of the country, geographically located near Armenia, Azerbaijan, Turkey and Turkmenistan where E. multilocularis is endemic [35].
In Iran, in contrast to the large amount of epidemiology data available for E. granulosus [3], only a few studies have been performed on E. multilocularis. Echinococcus multilocularis was reported for the first time in 1971 in three of 30 red foxes (10%) in the Moghan Plain of north-western Iran [31]. Later, in 1992, a study in the Ardabil Province in northwestern Iran has shown that 22.9% of the red foxes and 16% of the jackals studied were infected with E. multilocularis [32]. The results of both studies were based on morphological identification of adult worms. In a subsequent study in 2009 on canine echinococcosis in Moghan Plain, using copro-PCR and CA-ELISA, E. multilocularis was not reported [51]. In a recent study, E. multilocularis was reported in carnivores in Chenaran City, Razavi Khorasan Province in the north-east of the country for the first time, using multiplex PCR targeting nad1 on DNA extracted from the stool samples of the hosts [33]. The present study was undertaken due to a lack of knowledge on the distribution of echinococcosis in definitive hosts in North-Khorasan Province, which is located on the expanded distributional range of E. multilocularis from the northwest to the northeast of the country. To our knowledge, this is the first study on E. multilocularis in Iran utilizing both morphological characteristics of rostellar hooks and strobila of adult worms as well as molecular analysis of two genomic regions, cox1 and nad1.
Overall, morphological descriptions of adult Echinococcus species and molecular identification of both mitochondrial genes were in agreement. For species identification based on light microscopy, the dimensions and shape of the rostellar large and small hooks were useful. With regard to the strobila, the position of the genital pores of proglottids and the shape of the uterus in the gravid segments were important distinguishing factors between the two Echinococcus species. However, in immature worms, there was an overlap in the size of strobila and proglottids of E. granulosus and E. multilocularis, which needs consideration during morphological analysis of adults of these two species to prevent misidentification.
Overall, 17 animals out of 106 examined (16%) had echinococcosis. The overall rates of infection with E. multilocularis and E. granulosus were 9.4% and 6.6%, respectively. In jackals, both E. multilocularis (13.1%) and E. granulosus (3.3%) were found. Infection of this canid with these species of Echinococcus has been reported on multiple occasions and across a wide geographical range [52]. In Iran, infections in jackals with E. multilocularis has been reported from Ardabil Province in the northwest [32] and Razavi Khorasan Province in the northeast of Iran [33]. In Hungary, the jackal was recently reported as a new host record for E. multilocularis [53]. The jackal is under significant and fast geographical expansion [52] and can migrate long distances through ecological corridors [53]. Since this species is an important definitive host for echinococcosis, it can serve as a potential source of infection to humans and domestic animals in endemic areas.
The other canids studied here were infected with either E. multilocularis or E. granulosus. In foxes, the overall infection rate of E. multilocularis was 8.7%, and in dogs, the overall infection rate with E. granulosus was 21%. Among the three wolves available for examination, one harboured E. granulosus. Among different canids examined in the Ardabil Province, northwest Iran [32], foxes were found infected with E. multilocularis while dogs and wolves harboured E. granulosus; these observations are in agreement with our findings. In Kazakhstan, which has one of the world’s largest wolf populations [54], 19.5% of evaluated wolves were observed to be infected with E. granulosus. Globally, E. multilocularis is widely prevalent among foxes [55,56,57,58] while E. granulosus appears much less abundant [59,60,61,62].
In dogs, although some studies reported the occurrence of E. multilocularis, the susceptibility of dogs to this species is estimated to be very low [3]. In the present study, the lack of E. granulosus in foxes and E. multilocularis in dogs and wolves may be due to the low sample sizes. Nevertheless, there seems to be ample evidence that foxes are more susceptible to E. multilocularis than E. granulosus, while for dogs and wolves it is vice versa.
In the present study, although all infected animals were males (17 out of 83) and none of the 23 female animals were infected, no statistical analysis was performed due to sex imbalance in the study sample. However, risk factors evaluation of echinococcosis in a highly endemic region of the Tibetan Plateau [63] indicated that male dogs were more likely to be infected than female dogs, based on univariate and multivariate logistic regression analysis (P < 0.01). This increased risk for male animals was attributed to the activity of maintaining territory and hunting, which is likely higher in male dogs compared with female dogs [63].
Molecular analyses of echinococcosis in Iran have been performed mostly on CE in humans and domestic animals [15, 19, 20, 22] and only to a limited extent on adult worms in the definitive hosts [24,25,26,27]. This difference mostly reflects difficulties related to field studies of definitive hosts, such as obtaining stray dogs and wild canids, contamination with viral infections (i.e. rabies) and high risk of hydatid cyst infection during examinations. Thus, the number of molecular studies on adult worms of E. granulosus in Iran is limited. To date, E. granulosus (s.s.) (G1–G3) and E. intermedius (G6) have been reported from Iranian canids. The first study in this respect appeared in 2012 in Lorestan Province, West of Iran, and involved genotyping 20 isolates of E. granulosus from dogs using sequencing of mitochondrial cox1 and nad1 genes. In that study, G1, G2 and G3 genotypes were reported [24]. In another survey in northwestern Iran, using the cox1 gene as a molecular marker, G1, G3 and G6 genotypes of E. granulosus were identified among 16 dogs [25]. Later, E. granulosus G1 and G3 genotypes were identified in dogs and G1 in jackals from the Caspian Sea, north of the country, using sequencing of the cox1 gene [26]. In the present study, which used both cox1 and nad1 genes, all seven E. granulosus isolates form canids of North Khorasan Province belonged to the G1 genotype. This is compatible with the results of all previous relevant studies in Iran and emphasizes G1 as the dominant genotype.
In the present study, the cox1 gene revealed more genetic diversity within the E. granulosus than within the E. multilocularis isolates. However, the nad1 gene showed a higher degree of sequence variation in E. multilocularis compared with E. granulosus isolates. Four representative haplotypes of this study along with reference genotypes/species of Echinococcus were included in the phylogenetic tree using Bayesian inference method. The phylogenetic tree indicated six different clusters for Echinococcus spp. As expected, G4 (E. equinus) and E. shiquicus were placed as two distinct clades. Two representative haplotypes of E. multilocularis isolates from our study (E.mkh2 and E.mkh4) and two geographical genotypes of E. multilocularis namely M1 (China, Alaska, North America) and M2 (European) formed a clade sister to E. shiquicus with maximum statistical support (pp = 1.00) [12, 13]. The distinct placement of E. multilocularis relative to E. shiquicus was poorly supported (pp = 0.50) but it is in concordance with some previous studies [19, 24, 64]. Echinococcus vogeli and E. oligarthrus formed a clade with high statistical support (pp = 1.00), which is in agreement with previous studies [7, 24, 64]. Echinococcus felidis was recovered as a sister taxon to G1–G3 genotypes in a distinct clade with maximum statistical support (pp = 1.00); this is in concordance with the studies of Lavikainen et al. [65], Hüttner et al. [66], Saarma et al. [7] and previous studies in Iran [15, 64]. Two E. granulosus haplotypes from the present study (E.gkh63 and E.gkh75) identified as the G1 genotype were grouped with reference G1 genotype within the cluster with G1–G3 genotypes with maximum statistical support (pp = 1.00). Our finding provides further evidence that G1–G3 genotypes are separate from other species or genotypes of Echinococcus and should be considered as E. granulosus (s.s.) [66, 67]. Furthermore, our data are in agreement with the statement that G2 is not a distinct genotype [8, 19, 68]. G6–G10 genotypes clustered separately from the G5 genotype within a clade with strong statistical support (pp = 1.00), which is in agreement with previous studies in Iran [15, 17, 26] and confirms reconstruction of the G5 genotype as E. ortleppi [11, 66, 67].
Conclusions
In conclusion, our study confirmed that both E. multilocularis and E. granulosus are present in canines of the North-Khorasan Province, Iran. The distribution of E. multilocularis is wider than previously known. The jackal acts as a definitive host for both E. multilocularis and E. granulosus, but the infection rate with the former species is higher. This poses a potentially large risk of AE transmission to humans, especially in rural areas where jackals roam closer to human settlements than other wild canids. The increasing public health concern of this lethal zoonotic disease requires surveillance and early diagnosis of the infection in at-risk populations in the country. Future studies aiming to identify intermediate hosts for E. multilocularis in this endemic area of echinococcosis are needed. In addition, this area is suitable for further studies on the comparison of the biology of E. granulosus and E. multilocularis, with regard to infectivity across domestic and wild hosts.
Availability of data and materials
The data supporting the conclusions of this article are included within the article. The accession numbers of the sequences deposited in GenBank are provided in Table 3.
Abbreviations
- AE:
-
alveolar echinococcosis
- CE:
-
cystic echinococcosis
- cox1:
-
cytochrome c oxidase subunit 1 gene
- IST:
-
intestinal scraping technique
- nad1:
-
NADH dehydrogenase subunit 1 gene
- VDRC:
-
Vector-Borne Diseases Research Center
References
Alvarez Rojas CA, Romig T, Lightowlers MW. Echinococcus granulosus sensu lato genotypes infecting humans—review of current knowledge. Int J Parasitol. 2014;44:9–18.
Eckert J, Gemmell M, Meslin F-X, Pawlowski Z. WHO-OIE Manual on echinococcosis in humans and animals: a public health problem of global concern. Paris: World Organisation for Animal Health; 2001.
Deplazes P, Rinaldi L, Rojas CA, Torgerson P, Harandi M, Romig T, et al. Global distribution of alveolar and cystic echinococcosis. Adv Parasitol. 2017;95:315–493.
Eckert J, Deplazes P. Biological, epidemiological, and clinical aspects of echinococcosis, a zoonosis of increasing concern. Clin Microbiol Rev. 2004;17:107–35.
Moro P, Schantz PM. Echinococcosis: a review. Int J Infect Dis. 2009;13:125–33.
Thompson RC, Lymbery AJ. The nature, extent and significance of variation within the genus Echinococcus. Adv Parasitol. 1988;27:209–58.
Saarma U, Jõgisalu I, Moks E, Varcasia A, Lavikainen A, Oksanen A, et al. A novel phylogeny for the genus Echinococcus, based on nuclear data, challenges relationships based on mitochondrial evidence. Parasitology. 2009;136:317–28.
Kinkar L, Laurimäe T, Sharbatkhori M, Mirhendi H, Kia EB, Ponce-Gordo F, et al. New mitogenome and nuclear evidence on the phylogeny and taxonomy of the highly zoonotic tapeworm Echinococcus granulosus sensu stricto. Infect Genet Evol. 2017;52:52–8.
Laurimäe T, Kinkar L, Moks E, Romig T, Omer RA, Casulli A, et al. Molecular phylogeny based on six nuclear genes suggests that Echinococcus granulosus sensu lato genotypes G6/G7 and G8/G10 can be regarded as two distinct species. Parasitology. 2018;12:1–9.
Thompson RCA. The taxonomy, phylogeny and transmission of Echinococcus. Exp Parasitol. 2008;119:439–46.
Lopez-Neyra CR, Soler Planas MA. Revision del genero Echinococcus Rudy description de una especie nuéva parasita intestinal del porro en Almeria. Rev Iber Parasitol. 1943;3:169–94.
Bowles J, Blair D, McManus DP. Genetic variants within the genus Echinococcus identified by mitochondrial DNA sequencing. Mol Biochem Parasitol. 1992;54:165–73.
Bowles J, McManus D. NADH dehydrogenase 1 gene sequences compared for species and strains of the genus Echinococcus. Int J Parasitol. 1993;23:969–72.
Fadakar B, Tabatabaei N, Borji H, Naghibi A. Genotyping of Echinococcus granulosus from goats and sheep indicating G7 genotype in goats in the Northeast of Iran. Vet Parasitol. 2015;214:204–7.
Sharbatkhori M, Tanzifi A, Rostami S, Rostami M, Fasihi Harandi M. Echinococcus granulosus sensu lato genotypes in domestic livestock and humans in Golestan Province, Iran. Rev Inst Med Trop Sao Paulo. 2016;58:38.
Ebrahimipour M, Sadjjadi SM, Yousofi Darani H, Najjari M. Molecular studies on cystic echinococcosis of camel (Camelus dromedarius) and report of Echinococcus ortleppi in Iran. Iran J Parasitol. 2017;12:323–31.
Pezeshki A, Akhlaghi L, Sharbatkhori M, Razmjou E, Oormazdi H, Mohebali M, et al. Genotyping of Echinococcus granulosus from domestic animals and humans from Ardabil Province, northwest Iran. J Helminthol. 2013;87:387–91.
Eslami A, Shayan P, Bokaei S. Morphological and genetic characteristics of the liver hydatid cyst of a donkey with Iran origin. Iran J Parasitol. 2014;9:302–10.
Sharbatkhori M, Mirhendi H, Jex AR, Pangasa A, Campbell BE, Kia EB, et al. Genetic categorization of Echinococcus granulosus from humans and herbivorous hosts in Iran using an integrated mutation scanning-phylogenetic approach. Electrophoresis. 2009;30:2648–55.
Kia EB, Rahimi H, Sharbatkhori M, Talebi A, Harandi MF, Mirhendi H. Genotype identification of human cystic echinococcosis in Isfahan, central Iran. Parasitol Res. 2010;107:757–60.
Sadjjadi S, Mikaeili F, Karamian M, Maraghi S, Sadjjadi F, Shariat-Torbaghan S, et al. Evidence that the Echinococcus granulosus G6 genotype has an affinity for the brain in humans. Int J Parasitol. 2013;43:875–7.
Rostami S, Shariat Torbaghan S, Dabiri S, Babaei Z, Ali Mohammadi M, Sharbatkhori M, et al. Genetic characterization of Echinococcus granulosus from a large number of formalin-fixed, paraffin-embedded tissue samples of human isolates in Iran. Am J Trop Med Hyg. 2015;92:588–94.
Nikmanesh B, Mirhendi H, Ghalavand Z, Alebouyeh M, Sharbatkhori M, Kia EB, et al. Genotyping of Echinococcus granulosus isolates from human clinical samples based on sequencing of mitochondrial genes in Iran, Tehran. Iran J Parasitol. 2014;9:20–7.
Parsa F, Fasihi Harandi M, Rostami S, Sharbatkhori M. Genotyping Echinococcus granulosus from dogs from western Iran. Exp Parasitol. 2012;132:308–12.
Shariatzadeh SA, Spotin A, Gholami S, Fallah E, Hazratian T, Mahami-Oskouei M, et al. The first morphometric and phylogenetic perspective on molecular epidemiology of Echinococcus granulosus sensu lato in stray dogs in a hyperendemic Middle East focus, northwestern Iran. Parasit Vectors. 2015;8:409.
Gholami S, Jahandar H, Abastabar M, Pagheh A, Mobedi I, Sharbatkhori M. Echinococcus granulosus sensu stricto in dogs and jackals from Caspian Sea region, northern Iran. Iran J Parasitol. 2016;11:186.
Ghabdian S, Borji H, Naghibi A. Molecular identification of Echinococcus granulosus strain in stray dogs from northeastern Iran. Vet Parasitol Reg Stud Reports. 2017;9:6–8.
Fattahi Masoom S, Sharifi N. Alveolar hydatid disease: a case report. Med J Islam Repub Iran. 2007;20:205–7.
Geramizadeh B, Nikeghbalian S, Malekhosseini SA. Alveolar echinococcosis of the liver: report of three cases from different geographic areas of Iran. Hepat Mon. 2012;12:1–4.
Mobedi I, Dalimi A. Epidemiology of hydatid cyst in Iran and world (in Persian). Tehran: Moghaddam Publication; 1994. p. 132–47.
Mobedi I, Sadighian A. Echinococcus multilocularis Leuckart, 1863, in red foxes, Vulpes vulpes Linn., in Moghan, Azerbaijan Province, northwest of Iran. J Parasitol. 1971;57:493.
Zariffard M, Massoud J. Study of Echinococcus granulosus and Echinococcus multilocularis infections in Canidiae in Ardabile Province of Iran. Arch Razi Inst. 1998;48(49):47–52.
Beiromvand M, Akhlaghi L, Massom SHF, Mobedi I, Meamar AR, Oormazdi H, et al. Detection of Echinococcus multilocularis in carnivores in Razavi Khorasan Province, Iran using mitochondrial DNA. PLoS Negl Trop Dis. 2011;5:e1379.
Sadjjadi SM. Present situation of echinococcosis in the Middle East and Arabic North Africa. Parasitol Int. 2006;55:S197–202.
Torgerson PR, Keller K, Magnotta M, Ragland N. The global burden of alveolar echinococcosis. PLoS Negl Trop Dis. 2010;4:e722.
Zarei M, Talebzadeh H, Keyvan M. Report of a rare case of multiple hydatid cysts and perforation of a large abdominal cyst in a patient. Nursing. 2015;4:302–3.
Salehi M, Adinezade A, Khodajo R, Saberi Z, Yousefi A. The epidemiologic survey of operated patients with hydatid cyst in hospitals of North Khorasan Province during 2010–2011 (in Persian). J North Khorasan Univ Med Sci. 2013;4:623–9.
Shahkaram R, Sabzevari S. The simultaneous presence of hydatid cysts in the liver and spleen: a case report with splenectomy. J Biomed Health. 2018;3:24–30.
Takeuchi-Storm N, Woolsey ID, Jensen PM, Fredensborg BL, Pipper CB, Kapel CMO. Predictors of Echinococcus multilocularis prevalence in definitive and intermediate hosts: a meta-analysis approach. J Parasitol. 2015;101:297–303.
Darvish J, Siahsarvie R, Mirshamsi Kakhki O, Kayvanfar N, Hashemi N, Sadeghie Shakib F. Diversity of the rodents of northeastern Iran. Iran J Anim Biosyst. 2006;2:57–76.
Mohebali M, Arzamani K, Zarei Z, Akhoundi B, Hajjaran H, Raeghi S, et al. Canine visceral leishmaniasis in wild canines (fox, jackal, and wolf) in northeastern Iran using parasitological, serological, and molecular methods. J Arthropod Borne Dis. 2016;10:538–45.
Deplazes P, Eckert J. Diagnosis of the Echinococcus multilocularis infection in final hosts. Appl Parasitol. 1996;37:245–52.
Oryan A, Nazifi S, Sharifiyazdi H, Ahmadnia S. Pathological, molecular, and biochemical characterization of Coenurus gaigeri in Iranian native goats. J Parasitol. 2010;96:961–8.
Ash LR, Orihel TC. Parasites: a guide to laboratory procedures and identification. Chicago: American Society of Clinical Pathologists Press; 1991.
Soulsby E. Helminths, arthropods and protozoa of domesticated animals (sixth edition of MSnnigʼs Veterinary helminthology and entomology). London: Baillière, Tindall & Cassell; 1968.
Khalil LF, Jones A, Bray RA. Keys to the cestode parasites of vertebrates. Wallingford: CAB International; 1994.
Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–8.
Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinform Appl Note. 2001;17:754–5.
Page RD. TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–8.
Hotez PJ, Savioli L, Fenwick A. Neglected tropical diseases of the Middle East and North Africa: review of their prevalence, distribution, and opportunities for Control. PLoS Negl Trop Dis. 2012;6:e1475.
Zare-Bidaki M, Mobedi I, Naddaf S, Kia E, Mahmoudi M, Piazak N, et al. Prevalence of Echinococcus spp. infection using coproantigen ELISA among canids of Moghan Plain, Iran. Iran J Public Health. 2009;38:112–8.
Gherman CM, Mihalca AD. A synoptic overview of golden jackal parasites reveals high diversity of species. Parasit Vectors. 2017;10:419.
Széll Z, Marucci G, Pozio E, Sréter T. Echinococcus multilocularis and Trichinella spiralis in golden jackals (Canis aureus) of Hungary. Vet Parasitol. 2013;197:393–6.
Abdybekova AM, Torgerson PR. Frequency distributions of helminths of wolves in Kazakhstan. Vet Parasitol. 2012;184:348–51.
Deplazes P, Dinkel A, Mathis A. Molecular tools for studies on the transmission biology of Echinococcus multilocularis. Parasitology. 2003;127(Suppl):S53–61.
Nonaka N, Tsukada H, Abe N, Oku Y, Kamiya M. Monitoring of Echinococcus multilocularis infection in red foxes in Shiretoko, Japan, by coproantigen detection. Parasitology. 1998;117:193–200.
Sikó SB, Deplazes P, Ceica C, Tivadar C, Bogolin I, Popescu S, et al. Echinococcus multilocularis in south-eastern Europe (Romania). Parasitol Res. 2011;108:1093–7.
Bagrade G, Deksne G, Ozoliņa Z, Howlett SJ, Interisano M, Casulli A, Pozio E. Echinococcus multilocularis in foxes and raccoon dogs: an increasing concern for Baltic countries. Parasit Vectors. 2016;9:615.
Scioscia NP, Petrigh RS, Beldomenico PM, Fugassa M, Denegri GM. Survey and first molecular characterization of Echinococcus granulosus sensu stricto (G1) in Pampas fox (Lycalopex gymnocercus) in Buenos Aires Province, Argentina. Acta Trop. 2016;158:1–5.
Craig PS, Woods ML, Boufana B, O’Loughlin B, Gimpel J, San Lett W, et al. Cystic echinococcosis in a fox-hound hunt worker, UK. Pathog Glob Health. 2012;106:373–5.
Lahmar S, Boufana B, Boubaker SB, Landolsi F. Intestinal helminths of golden jackals and red foxes from Tunisia. Vet Parasitol. 2014;204:297–303.
Acosta-Jamett G, Cleaveland S, de Bronsvoort B, Cunningham A, Bradshaw H, Craig P. Echinococcus granulosus infection in foxes in Coquimbo District, Chile. Arch Med Vet. 2015;47:409–13.
Budke CM, Campos-Ponce M, Qian W, Torgerson PR. A canine purgation study and risk factor analysis for echinococcosis in a high endemic region of the Tibetan plateau. Vet Parasitol. 2005;127:43–9.
Hajialilo E, Harandi MF, Sharbatkhori M, Mirhendi H, Rostami S. Genetic characterization of Echinococcus granulosus in camels, cattle and sheep from the south-east of Iran indicates the presence of the G3 genotype. J Helminthol. 2012;86:263–70.
Lavikainen A, Haukisalmi V, Lehtinen MJ, Henttonen H, Oksanen A, Meri S. A phylogeny of members of the family Taeniidae based on the mitochondrial cox1 and nad1 gene data. Parasitology. 2008;135:1457–67.
Hüttner M, Nakao M, Wassermann T, Siefert L, Boomker JD, Dinkel A, et al. Genetic characterization and phylogenetic position of Echinococcus felidis (Cestoda: Taeniidae) from the African lion. Int J Parasitol. 2008;38:861–8.
Nakao M, McManus DP, Schantz PM, Craig PS, Ito A. A molecular phylogeny of the genus Echinococcus inferred from complete mitochondrial genomes. Parasitology. 2007;134:713–22.
Kinkar L, Laurimäe T, Acosta-Jamett G, Andresiuk V, Balkaya I, Casulli A, et al. Distinguishing Echinococcus granulosus sensu stricto genotypes G1 and G3 with confidence: a practical guide. Infect Genet Evol. 2018;64:178–84.
Le T, Pearson M, Blair D, Dai N, Zhang L, McManus D. Complete mitochondrial genomes confirm the distinctiveness of the horse-dog and sheep-dog strains of Echinococcus granulosus. Parasitology. 2002;124:97–112.
Varcasia A, Canu S, Kogkos A, Pipia AP, Scala A, Garippa G, et al. Molecular characterization of Echinococcus granulosus in sheep and goats of Peloponnesus, Greece. Parasitol Res. 2007;101:1135–9.
Lavikainen A, Lehtinen M, Meri T, Hirvelä-Koski V, Meri S. Molecular genetic characterization of the Fennoscandian cervid strain, a new genotypic group (G10) of Echinococcus granulosus. Parasitology. 2003;127:207–15.
Bowles J, Blair D, McManus D. Molecular genetic characterization of the cervid strain (‘northern form’) of Echinococcus granulosus. Parasitology. 1994;109:215–21.
Gasser RB, Zhu X, McManus DP. NADH dehydrogenase subunit 1 and cytochrome c oxidase subunit I sequences compared for members of the genus Taenia (Cestoda). Int J Parasitol. 1999;29:1965–70.
Acknowledgements
Thanks to professor Peter Deplazes (University of Zurich, Switzerland) for his valuable advice. The authors also would like to thank Dr Ahmad Moosazadeh, Mr Mahmoud Yazdani, Mr Javad Namjou and Mr Nima Esmati from the Vector-Borne Diseases Research Center, North Khorasan University of Medical Sciences, Iran, for field assistance.
Funding
This study was part of the Ph.D. Thesis of first author (ZH) in the Department of Medical Parasitology and Mycology, School of Public Health, Tehran University of Medical Sciences, Iran. The study was approved and financially supported by the Deputy of Research, Tehran University of Medical Sciences, Tehran, Iran, through Grant No. 92-02-160-23365.
Author information
Authors and Affiliations
Contributions
ZH, MM and EBK designed the study. ZZ and KA carried out field activities, collected the samples and prepared them for laboratory processing. ZH performed laboratory experiments. IM participated in morphological species identification of Echinococcus. ZH, MiS, SHM, BN and MeS contributed to analysis of DNA sequence variations of Echinococcus. ZH, MM, MeS and EBK analyzed the data. ZH and MiS wrote the draft of the manuscript. EBK supervised the project and finalized the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Heidari, Z., Sharbatkhori, M., Mobedi, I. et al. Echinococcus multilocularis and Echinococcus granulosus in canines in North-Khorasan Province, northeastern Iran, identified using morphology and genetic characterization of mitochondrial DNA. Parasites Vectors 12, 606 (2019). https://doi.org/10.1186/s13071-019-3859-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-019-3859-z