Abstract
Background
Cryptosporidium spp. and Enterocytozoon bieneusi are prevalent zoonotic pathogens responsible for the high burden of diarrheal diseases worldwide. Rodents are globally overpopulated and are known as reservoirs or carriers of a variety of zoonotic pathogens including Cryptosporidium spp. and E. bieneusi. However, few data are available on genetic characterizations of both pathogens in rodents in China. The aim of the present work was to determine the prevalence and genetic characterizations of Cryptosporidium spp. and E. bieneusi in brown rats (Rattus norvegicus) from Heilongjiang, China.
Methods
A total of 242 wild brown rats were captured in Heilongjiang Province of China. A fresh fecal specimen was collected directly from the intestinal and rectal content of each brown rat. All the fecal specimens were examined for the presence of Cryptosporidium spp. and E. bieneusi by PCR and sequencing of the partial small subunit (SSU) rRNA gene and the internal transcribed spacer (ITS) region of the rRNA gene of the two pathogens, respectively.
Results
The infection rate was 9.1% (22/242) for Cryptosporidium spp. and 7.9% (19/242) for E. bieneusi. Sequence analysis confirmed the presence of C. ubiquitum (1/22, 4.5%) and three genotypes of Cryptosporidium, including Cryptosporidium rat genotype I (14/22, 63.6%), Cryptosporidium rat genotype IV (6/22, 27.3%) and Cryptosporidium suis-like genotype (1/22, 4.5%). Meanwhile, two E. bieneusi genotypes were identified, including D (17/19, 89.5%) and Peru6 (2/19, 10.5%).
Conclusions
To the best of our knowledge, Enterocytozoon bieneusi genotype Peru6 was identified in rodents for the first time globally and Cryptosporidium rat genotype I and Cryptosporidium rat genotype IV were found in rats in China for the first time. The finding of zoonotic C. ubiquitum and C. suis-like genotype, as well as E. bieneusi genotypes, suggests that brown rats pose a threat to human health. It is necessary to control brown rat population in the investigated areas and improve local people’s awareness of the transmission risk of the two pathogens from brown rats to humans.
Similar content being viewed by others
Background
Cryptosporidium spp. and Enterocytozoon bieneusi are two common opportunistic pathogens in humans and have been reported to be associated with diarrhea [1, 2]. However, clinical symptoms of the infection are variable depending on the health status of the infected hosts, displaying asymptomatic infection or self-limiting diarrhea in healthy people, and chronic or life-threatening diarrhea in immunocompromised individuals [2, 3]. In addition to humans, numerous animal species are also the hosts of the two pathogens, suggesting a zoonotic nature of the two parasitic diseases [4, 5]. Meanwhile, they have also been identified in some water bodies and food products, indicating the possibility of water-borne and food-borne transmission [3, 6, 7]. Because of the clinical and public health importance of Cryptosporidium spp. and E. bieneusi, both have been ranked on category B list, in which the pathogens are defined as the second highest priority organisms/biological agents by the National Institutes of Health (NIH) of the USA [8]. Meanwhile, Cryptosporidium spp. is ranked fifth among the 24 most important food-borne parasites in a global ranking by a joint Food and Agriculture Organization (FAO)/World Health Organization (WHO) expert committee [9]. Enterocytozoon bieneusi is listed on the Environmental Protection Agency (EPA) microbial contaminant candidate list of concern for waterborne transmission [10].
Cryptosporidium is a complex genus. To date, at least 31 species and more than 40 genotypes of Cryptosporidium have been identified by sequencing the small subunit (SSU) rRNA gene [2, 11]. Of them, 21 Cryptosporidium species/genotypes have been isolated in humans and C. parvum is generally considered to be zoonotically transmitted [2]. Some zoonotic outbreaks of cryptosporidiosis caused by C. parvum have been reported and confirmed at a subtype level, such as calf-derived IIaA15G2R1 in the UK [12] and sheep-derived IIaA17G1R1, IIaA15G2R1 in the UK and IIaA20G2R1 in Italy [12, 13]. Thus, molecular epidemiological investigations of animal cryptosporidiosis have been paid more attention. However, most of them focus on farm animals (pigs, sheep and cattle) or pets (dogs and cats) [5, 14]. Rodents, as the most widespread groups of mammals, have been reported to carry at least 11 species and more than 20 genotypes of Cryptosporidium as vectors or reservoirs, including C. parvum, C. muris, C. ubiquitum, C. meleagridis, C. scrofarum, C. wrairi, C. tyzzeri, C. rubeyi, C. andersoni, C. hominis, C. suis and rat genotypes (I-IV), mouse genotypes (II, III) and the Naruko genotype, ferret genotype, chipmunk genotypes (I, II), skunk genotype, hamster genotype, deer mouse genotypes (I-IV), vole genotype, bear genotype, muskrat genotypes (I, II) and ground squirrel genotypes (I-III) [15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38]. Among them, Cryptosporidium species except for C. rubeyi and two genotypes (chipmunk genotype I and skunk genotype) have also been found in humans [11].
For E. bieneusi, at least 240 genotypes have been identified by analyzing the internal transcribed spacer (ITS) region (~243 bp) of the rRNA gene and they have been classified into nine groups (groups 1-9) by phylogenetic analysis [39, 40]. Group 1 is composed of the common zoonotic genotypes and groups 2-9 mostly contain host-adapted genotypes [11]. To date, more than 70 genotypes have been found in humans, 33 of which are also found in animals, supporting presumption of zoonotic potential [1, 5]. In fact, zoonotic transmission of E. bieneusi has been reported in Peru, which occurred between a child and guinea pigs [41]. Enterocytozoon bieneusi has been detected in many rodent species and 35 genotypes have been identified, including 12 zoonotic genotypes (BEB6, C, D, EbpA, EbpC, H, Peru8, Peru11, Peru16, PigITS5, S6 and TypeIV) [22, 41,42,43,44,45,46,47,48].
Currently, due to limited effect of nitazoxanide on cryptosporidiosis and fumagillin on microsporidiosis caused by E. bieneusi [11], transmission control and prevention of infection of the two pathogens targeting the epidemiologic cycles are the key effective strategies. Understanding Cryptosporidium spp. and E. bieneusi epidemiology in wide range of hosts, exploring the molecular phylogeny and assessing zoonotic potential of animal-derived isolates are the key steps to prevent and reduce occurrence of the two parasitic diseases. Brown rats (Rattus norvegicus) are one of the most common rodent species, and usually live almost everywhere humans are. The dynamic activity of rodents facilitates the transmission and spread of various diseases including cryptosporidiosis and microsporidiosis caused by E. bieneusi [49].
In Heilongjiang Province of China, Cryptosporidium spp. and E. bieneusi are prevalent in a variety of species of animals [50, 51]. Moreover, they have been found in human immunodeficiency virus (HIV)-infected and acquired immunodeficiency syndrome (AIDS)-patients (unpublished data), cancer patients and children [52, 53]. The source of the human infection is still unclear. This study aimed to determine the prevalence of Cryptosporidium spp. and E. bieneusi in brown rats from various regions of Heilongjiang, China, to characterize the isolates and assess their zoonotic potential.
Methods
Study sites and rodent collections
During a three-year period from April 2014 to June 2017, 242 brown rats were captured from five distinct regions of Heilongjiang Province, China, including 55 from a granary in Xingren Town, 30 from a cattle farm in Xingren Town, 73 from a pig farm in Mingshui County, 27 from a pig farm in Qinggang County, 37 from a sheep farm in Baoqing County and 20 from a subdistrict in Harbin City (Table 1). All rats were captured in cage traps baited with sunflower seeds and peanut/sesame butter. In each location, 20 cage traps were installed at sunset and gathered before sunrise, with traps 5 m apart in transects. All rats were transported to the laboratory within 48 h after being captured and were killed by CO2 inhalation.
Fecal sample collection and DNA extraction
A fresh fecal specimen (approximately 500 mg) was collected directly from the intestinal and rectal content of each brown rat. All specimens were washed with distilled water by centrifugation for 10 min at 1,500× g at room temperature. Genomic DNA was extracted directly from approximately 200 mg of each processed specimen using QIAamp DNA Mini Stool Kit (Qiagen, Hilden, Germany) according to the manufacturer’s procedures. The lysis temperature was increased to 95 °C in order to obtain high DNA yield. DNA was eluted in 200 μl of AE elution buffer (provided with the kit) and stored at -20 °C prior to PCR analysis.
Genotyping of Cryptosporidium spp. and E. bieneusi
Cryptosporidium spp. in the fecal specimens was identified by nested PCR amplification of a SSU rRNA gene fragment of ~830 bp designed by Xiao et al. [54]. Each PCR consisted of 35 cycles of denaturation at 94 °C for 45 s, annealing at 60 °C for 45 s, and extension at 72 °C for 60 s; an initial denaturation step consisting of incubation at 94 °C for 5 min and a final extension step consisting of incubation at 72 °C for 10 min were also included [54]. Enterocytozoon bieneusi was identified and genotyped by nested PCR amplification of an approximately 390 bp nucleotide fragment of the rRNA gene, containing 76 bp of the 3' end of the SSU rRNA gene, 243 bp of the ITS region, and 70 bp of the 5' end of the large subunit (LSU) rRNA gene designed by Buckholt et al. [55]. The two sets of cycling parameters were as follows: 35 cycles of 94 °C for 30 s, 57 °C for 30 s, and 72 °C for 40 s and 30 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 40 s, with both of them having an initial denaturation step at 94 °C for 5 min and a final extension step at 72 °C for 10 min [55]. TaKaRa Taq DNA polymerase (TaKaRa Bio Inc., Tokyo, Japan) was used for all the PCR amplifications. All PCR amplification tests were carried out with positive controls (chicken-derived C. bailey DNA for Cryptosporidium spp. and deer-derived genotype BEB6 DNA for E. bieneusi) and negative controls which contained no DNA.
DNA sequencing and analysis
All nested PCR products were sequenced using the same PCR primers used for the secondary PCRs on an ABI PRISMTM 3730 DNA Analyzer (Applied Biosystems, Carlsbad, CA, USA), using a BigDye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems). The accuracy of the sequencing data was confirmed by sequencing of the PCR products in both directions. Further PCR products were sequenced for some DNA preparations, from which we obtained the sequences with single nucleotide substitutions, deletions or insertions compared to those published in GenBank. The species and genotypes of Cryptosporidium and E. bieneusi isolates were identified by aligning and analyzing the nucleotide sequences with each other and with reference sequences from GenBank using the Basic Local Alignment Search Tool (BLAST) and Clustal X 1.83 [56]. All the E. bieneusi genotypes were named based on 243 bp of the ITS region according to the established nomenclature system [57].
Results
Prevalence of Cryptosporidium spp. and E. bieneusi
Totals of 22 (9.1%) and 19 (7.9%) out of 242 brown rats were found to be infected with Cryptosporidium spp. and E. bieneusi, respectively. Cryptosporidium spp. was found in five areas, with infection rates ranging between 6.7–16.4% except the subdistrict of Harbin City. Enterocytozoon bieneusi was found in all the six areas investigated, with infection rates ranging between 1.8–27.0%. No mixed infections of the two pathogens were found in rodents in our study (Table 1).
Cryptosporidium species/genotypes
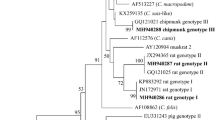
Sequence analysis of SSU rRNA gene products of 22 Cryptosporidium isolates identified four Cryptosporidium species and genotypes, including Cryptosporidium rat genotype I (14/22, 63.6%), Cryptosporidium rat genotype IV (6/22, 27.3%), Cryptosporidium suis-like genotype (1/22, 4.5%) and C. ubiquitum (1/22, 4.5%). Cryptosporidium rat genotype I showed dominance in brown rats in the investigated regions (Table 1).
At the SSU rRNA locus, 14 Cryptosporidium rat genotype I isolates had 100% homology between each other and were identical to that (FJ205699) from waste water in China and those from R. norvegicus in Sweden (JN172971), R. rattus in Iran (KP883289) and raw water in the UK (GQ183517). Of the six sequences of Cryptosporidium rat genotype IV, five have not been reported previously. Among them, four sequences were identical to each other (MG917670), with one base substitution at position 309 (C→T) when compared with that from raw water in the UK (GQ183515); the fifth (MG917671) had one base difference compared with that from storm water in the USA (AY737583). The sixth sequence had 100% similarity with those from R. norvegicus in Sweden (JN172970) and storm water in the USA (AY737585). The sequences of C. ubiquitum and Cryptosporidium suis-like isolates obtained here had 100% similarity with those from Apodemus flavicollis in Poland (KC962124) and a human in the UK (HQ822146), respectively.
Enterocytozoon bieneusi genotypes
By analyzing nucleotide sequence of the ITS region of the rRNA gene of E. bieneusi, we identified two known genotypes D and Peru6 in the brown rats, which had 100% homology with the two sequences GQ406055 and KX375800, respectively. Genotype D was found to be dominant, in 88.9% (17/19) of E. bieneusi isolates. Genotype D was also found to have a wide distribution, identified in all regions investigated. The remaining two rodents were infected with genotype Peru6, with 10.5% (2/19) frequency. This genotype was only found in two brown rats captured in the sheep farm (Table 1).
Discussion
In the present study, 9.1% of the brown rats examined were found to be infected with Cryptosporidium spp. The infection rate of Cryptosporidium was lower than those reported in brown rats in Iran (17.1%), Japan (38.0%), the Philippines (18.6%) and Sweden (12.0%) [23, 26, 31, 35], but higher than those reported in China (5.6% and 7.1%), Iran (4.1%) and Nigeria (1.5%) [19, 20, 24, 30]. Cryptosporidium spp. has been detected in various rodent species. Variable prevalence rates have been observed, such as 8.0–31.4% in mice, 2.1–63.0% in rats and 0.8–73.0% in voles [58], with the lowest and the highest prevalence rates in muskrats (0.7%) and in guinea pigs (85.0%), respectively [19, 59]. In the present study, E. bieneusi was detected in brown rats for the first time, with a prevalence of 7.9%. To date, there are only eight studies of E. bieneusi infection in rodents worldwide [22, 41,42,43,44,45,46,47]. In general, the prevalence of E. bieneusi in brown rats here was higher than those in chinchillas (3.6%) and mice (1.1%) [22, 45], but lower than those in hamster family (24.3%), prairie dogs (48.3%), squirrel family (16.7–42.9%), voles (39.1%) and mice (10.5–87.5%) [41,42,43,44, 46, 47]. The difference in prevalence may be related to rodent species, detection methods, sample size, animal age and study locations [58].
In the present study, four Cryptosporidium species/genotypes were identified including Cryptosporidium rat genotype I and IV, Cryptosporidium suis-like genotype and C. ubiquitum. Previous molecular epidemiological data revealed the presence of at least 11 species and 20 genotypes of Cryptosporidium spp. in rodents worldwide [15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38]. In China, five Cryptosporidium species (C. parvum, C. muris, C. andersoni, C. ubiquitum and C. wrairi) and six Cryptosporidium genotypes (mouse genotype I, rat genotypes II and III, ferret genotype, chipmunk genotype III and hamster genotype) have been found in rodents [19, 20, 22]. The species and genotype identification of rodent-derived Cryptosporidium spp. will be helpful to understand the roles that rodents play in the transmission of cryptosporidiosis.
Several studies have revealed that rats appear to be a major animal host for Cryptosporidium rat genotype I and Cryptosporidium rat genotype IV. Cryptosporidium rat genotype I (previously rat genotype) has been found in brown rats from Philippines, Sweden and Nigeria, and Cryptosporidium rat genotype IV (previously W19) in brown rats from Japan and Sweden [26, 30, 31, 35]. Furthermore, the two genotypes have also been detected in environmental samples, including a stream in the USA [48], raw water in the UK and China [60, 61] and the South Nation River watershed in Canada [62]. However, Cryptosporidium rat genotype I and Cryptosporidium rat genotype IV were found in brown rats in China for the first time. To date, the potential of the two genotypes causing disease in humans or livestock is unknown. In the future, more systematic molecular epidemiological investigations of Cryptosporidium spp. in more hosts need to be carried out to understand the true host range of the two genotypes.
Cryptosporidium ubiquitum, previously known as the Cryptosporidium cervine genotype, infects the largest number of host species of animals [63]. It has been found in domesticated and wild ruminants, a colony of non-human primates (lemurs) and a variety of rodents [22, 29, 32, 35, 38, 63]. To date, human cases of cryptosporidiosis caused by C. ubiquitum have been documented in more than ten countries [64]. In addition, this species has also been found in some water bodies, including source water, storm water and raw waste-water [63]. Cryptosporidium suis-like genotype has been recorded in rats in Philippines [31] and other animal species including cattle in Denmark, India and China, and yaks in China [65,66,67,68]. This genotype was also found in humans in Canada [69]. Although C. ubiquitum and Cryptosporidium suis-like genotype only accounted for 9.1% of all the Cryptosporidium isolates in investigated brown rats, we still need to consider them as a threat to human health, especially Cryptosporidium suis-like genotype, for this genotype was found in a granary.
To date, 35 E. bieneusi genotypes have been identified in rodents worldwide, 12 of which (BEB6, C, D, EbpA, EbpC, H, Peru 8, Peru11, Peru 16, PigITS5, S6 and TypeIV) have been detected in humans [22, 41,42,43,44,45,46,47,48]. In the present study, two known zoonotic E. bieneusi genotypes, D and Peru 6, were identified in brown rats. Genotype D was identified in 88.9% of E. bieneusi isolates and was found to have a wide distribution; it was found in all areas investigated. Genotype D is reported to be responsible for most human infections and it has been found in humans from more than 40 countries or areas [1]. It is also isolated in at least 15 species of animals as well as in some water bodies [70, 71]. In the present study Peru6 was only identified in two E. bieneusi isolates from two rats captured in the sheep farm. This genotype has been recorded in humans in Peru and Portugal [72,73,74], and some mammalian animal species and bird species [5, 75]. It has also been detected in wastewater in China [71]. To our knowledge, genotype Peru6 was found in rodents for the first time globally, indicating that this genotype might have more reservoir hosts than expected. The finding of two genotypes previously reported in humans suggests the possibility of rodents in the transmission of E. bieneusi to humans [1, 5].
Conclusions
The present study demonstrated the occurrence of Cryptosporidium spp. and E. bieneusi in brown rats in Heilongjiang, China and genetically characterized the isolates. We identified E. bieneusi genotype Peru6 in rodents for the first time, and Cryptosporidium rat genotype I and Cryptosporidium rat genotype IV in rats in China for the first time. The finding of zoonotic C. ubiquitum and Cryptosporidium suis-like genotype as well as two E. bieneusi genotypes suggests that brown rats pose a threat to human health. Thus, it is strongly recommended to take measures to control brown rat populations in the areas investigated and improve local people’s awareness of the transmission risk of these two diseases from brown rats to humans.
Abbreviations
- AIDS:
-
Acquired immunodeficiency syndrome
- BLAST:
-
Basic Local Alignment Search Tool
- EPA:
-
Environmental Protection Agency
- HIV:
-
Human immunodeficiency virus
- ITS:
-
Internal transcribed spacer
- NIH:
-
National Institutes of Health
- SSU:
-
Small subunit
References
Matos O, Lobo ML, Xiao L. Epidemiology of Enterocytozoon bieneusi infection in humans. J Parasit Res. 2012;2012:981424.
Ryan U, Fayer R, Xiao L. Cryptosporidium species in humans and animals: Current understanding and research needs. Parasitology. 2014;141:1667–85.
Didier ES, Weiss LM. Microsporidiosis: current status. Curr Opin Infect Dis. 2006;19:485–92.
Ryan U, Zahedi A, Paparini A. Cryptosporidium in humans and animals - a One Health approach to prophylaxis. Parasite Immunol. 2016;38:535–47.
Santín M, Fayer R. Microsporidiosis: Enterocytozoon bieneusi in domesticated and wild animals. Res Vet Sci. 2011;90:363–71.
Ryan U, Hijjawi N, Xiao L. Foodborne cryptosporidiosis. Int J Parasitol. 2018;48:1–12.
Speich B, Croll D, Fürst T, Utzinger J, Keiser J. Effect of sanitation and water treatment on intestinal protozoa infection: a systematic review and meta-analysis. Lancet Infect Dis. 2016;6:87–99.
NIAID Emerging Infectious Diseases/Pathogens. (https://www.niaid.nih.gov/research/emerging-infectious-diseases-pathogens). Accessed 7 May 2018.
Report of a Joint FAO/WHO Expert Meeting, 3–7 September 2012, FAO Headquarters, Rome, Italy. (http://www.fao.org/food/food-safety-quality/a-z-index/foodborne-parasites/en/). Accessed 7 May 2018.
Didier ES, Weiss LM, Cali A, Marciano-Cabral F. Overview of the presentations on microsporidia and free-living amebae at the 10th International Workshops on Opportunistic Protists. Eukaryot Cell. 2009;8:441–5.
Yang Z, Zhao W, Shen Y, Zhang W, Shi Y, Ren G, et al. Subtyping of Cryptosporidium cuniculus and genotyping of Enterocytozoon bieneusi in rabbits in two farms in Heilongjiang Province, China. Parasite. 2016;23:52.
Chalmers RM, Giles M. Zoonotic cryptosporidiosis in the UK - challenges for control. J Appl Microbiol. 2010;109:1487–97.
Cacciò SM, Sannella AR, Mariano V, Valentini S, Berti F, Tosini F, Pozio E. A rare Cryptosporidium parvum genotype associated with infection of lambs and zoonotic transmission in Italy. Vet Parasitol. 2013;191:128–31.
Feng Y, Xiao L. Molecular epidemiology of Cryptosporidiosis in China. Front Microbiol. 2017;8:1701.
Paparini A, Jackson B, Ward S, Young S, Ryan UM. Multiple Cryptosporidium genotypes detected in wild black rats (Rattus rattus) from northern Australia. Exp Parasitol. 2012;131:404–12.
Foo C, Farrell J, Boxell A, Robertson I, Ryan UM. Novel Cryptosporidium genotype in wild Australian mice (Mus domesticus). Appl Environ Microbiol. 2007;73:7693–6.
Warren KS, Swan RA, Morgan-Ryan UM, Friend JA, Elliot A. Cryptosporidium muris infection in bilbies (Macrotis lagotis). Aust Vet J. 2003;81:739–41.
Meireles MV, Soares RM, Bonello F, Gennari SM. Natural infection with zoonotic subtype of Cryptosporidium parvum in capybara (Hydrochoerus hydrochaeris) from Brazil. Vet Parasitol. 2007;147:166–70.
Lv C, Zhang L, Wang R, Jian F, Zhang S, Ning C, et al. Cryptosporidium spp. in wild, laboratory, and pet rodents in China: prevalence and molecular characterization. Appl Environ Microbiol. 2009;75:7692–9.
Zhao Z, Wang R, Zhao W, Qi M, Zhao J, Zhang L, et al. Genotyping and subtyping of Giardia and Cryptosporidium isolates from commensal rodents in China. Parasitology. 2015;142:800–6.
Li Q, Li L, Tao W, Jiang Y, Wan Q, Lin Y, et al. Molecular investigation of Cryptosporidium in small caged pets in northeast China: host specificity and zoonotic implications. Parasitol Res. 2016;115:2905–11.
Qi M, Luo N, Wang H, Yu F, Wang R, Huang J, et al. Zoonotic Cryptosporidium spp. and Enterocytozoon bieneusi in pet chinchillas (Chinchilla lanigera) in China. Parasitol Int. 2015;64:339–41.
Gholipoury M, Rezai HR, Namroodi S, Arab Khazaeli F. Zoonotic and non-zoonotic parasites of wild rodents in Turkman Sahra, northeastern Iran. Iran J Parasitol. 2016;11:350–7.
Saki J, Foroutan-Rad M, Asadpouri R. Molecular characterization of Cryptosporidium spp. in wild rodents of southwestern Iran using 18S rRNA gene nested-PCR-RFLP and sequencing techniques. J Trop Med. 2016;2016:6834206.
Kvác M, Hofmannová L, Bertolino S, Wauters L, Tosi G, Modrý D. Natural infection with two genotypes of Cryptosporidium in red squirrels (Sciurus vulgaris) in Italy. Folia Parasitol. 2008;55:95–9.
Kimura A, Edagawa A, Okada K, Takimoto A, Yonesho S, Karanis P. Detection and genotyping of Cryptosporidium from brown rats (Rattus norvegicus) captured in an urban area of Japan. Parasitol Res. 2007;100:1417–20.
Murakoshi F, Fukuda Y, Matsubara R, Kato Y, Sato R, Sasaki T, et al. Detection and genotyping of Cryptosporidium spp. in large Japanese field mice, Apodemus speciosus. Vet Parasitol. 2013;196:184–8.
Nakai Y, Hikosaka K, Sato M, Sasaki T, Kaneta Y, Okazaki N. Detection of Cryptosporidium muris type oocysts from beef cattle in a farm and from domestic and wild animals in and around the farm. J Vet Med Sci. 2004;66:983–4.
Song J, Kim CY, Chang SN, Abdelkader TS, Han J, Kim TH, et al. Detection and molecular characterization of Cryptosporidium spp. from wild rodents and insectivores in South Korea. Korean J Parasitol. 2015;53:737–43.
Ayinmode AB, Ogbonna NF, Widmer G. Detection and molecular identification of Cryptosporidium species in laboratory rats (Rattus norvegicus) in Ibadan, Nigeria. Ann Parasitol. 2017;63:105–9.
Ng-Hublin JS, Singleton GR, Ryan U. Molecular characterization of Cryptosporidium spp. from wild rats and mice from rural communities in the Philippines. Infect Genet Evol. 2013;16:5–12.
Matysiak A, Buńkowska-Gawlik K, Zaleśny G, Hildebrand J. Small rodents as reservoirs of Cryptosporidium spp. and Giardia spp. in south-western Poland. Ann Agric Environ Med. 2015;22:1–5.
Bajer A, Bednarska M, Pawelczyk A, Behnke JM, Gilbert FS, Sinski E. Prevalence and abundance of Cryptosporidium parvum and Giardia spp. in wild rural rodents from the Mazury Lake District region of Poland. Parasitology. 2002;125:21–34.
Danišová O, Valenčáková A, Stanko M, Luptáková L, Hatalová E, Čanády A. Rodents as a reservoir of infection caused by multiple zoonotic species/genotypes of C. parvum, C. hominis, C. suis, C. scrofarum, and the first evidence of Cryptosporidium muskrat genotypes I and II of rodents in Europe. Acta Trop. 2017;172:29–35.
Backhans A, Jacobson M, Hansson I, Lebbad M, Lambertz ST, Gammelgård E, et al. Occurrence of pathogens in wild rodents caught on Swedish pig and chicken farms. Epidemiol Infect. 2013;141:1885–91.
Stenger BLS, Clark ME, Kváč M, Khan E, Giddings CW, Prediger J, et al. North American tree squirrels and ground squirrels with overlapping ranges host different Cryptosporidium species and genotypes. Infect Genet Evol. 2015;36:287–93.
Stenger BL, Clark ME, Kváč M, Khan E, Giddings CW, Dyer NW, et al. Highly divergent 18S rRNA gene paralogs in a Cryptosporidium genotype from eastern chipmunks (Tamias striatus). Infect Genet Evol. 2015;32:113–23.
Feng Y, Alderisio KA, Yang W, Blancero LA, Kuhne WG, Nadareski CA, et al. Cryptosporidium genotypes in wildlife from a New York watershed. Appl Environ Microbiol. 2007;73:6475–83.
Karim MR, Dong H, Li T, Yu F, Li D, Zhang L, et al. Predomination and new genotypes of Enterocytozoon bieneusi in captive nonhuman primates in zoos in China: high genetic diversity and zoonotic significance. PLoS One. 2015;10:e0117991.
Zhao W, Zhang W, Yang Z, Liu A, Zhang L, Yang F, et al. Genotyping of Enterocytozoon bieneusi in farmed blue foxes (Alopex lagopus) and raccoon dogs (Nyctereutes procyonoides) in China. PLoS One. 2015;10:e0142611.
Cama VA, Pearson J, Cabrera L, Pacheco L, Gilman R, Meyer S, et al. Transmission of Enterocytozoon bieneusi between a child and guinea pigs. J Clin Microbiol. 2007;45:2708–10.
Deng L, Li W, Yu X, Gong C, Liu X, Zhong Z, et al. First report of the human-pathogenic Enterocytozoon bieneusi from red-bellied tree squirrels (Callosciurus erythraeus) in Sichuan, China. PLoS One. 2016;11:e0163605.
Sak B, Kváč M, Květoňová D, Albrecht T, Piálek J. The first report on natural Enterocytozoon bieneusi and Encephalitozoon spp. infections in wild East-European house mice (Mus musculus musculus) and West-European house mice (M. m. domesticus) in a hybrid zone across the Czech Republic-Germany border. Vet Parasitol. 2011;178:246–50.
Perec-Matysiak A, Buńkowska-Gawlik K, Kváč M, Sak B, Hildebrand J, Leśniańska K. Diversity of Enterocytozoon bieneusi genotypes among small rodents in southwestern Poland. Vet Parasitol. 2015;214:242–6.
Danišová O, Valenčáková A, Stanko M, Luptáková L, Hasajová A. First report of Enterocytozoon bieneusi and Encephalitozoon intestinalis infection of wild mice in Slovakia. Ann Agric Environ Med. 2015;22:251–2.
Guo Y, Alderisio KA, Yang W, Cama V, Feng Y, Xiao L. Host specificity and source of Enterocytozoon bieneusi genotypes in a drinking source watershed. Appl Environ Microbiol. 2014;80:218–25.
Roellig DM, Salzer JS, Carroll DS, Ritter JM, Drew C, Gallardo-Romero N, et al. Identification of Giardia duodenalis and Enterocytozoon bieneusi in an epizoological investigation of a laboratory colony of prairie dogs, Cynomys ludovicianus. Vet Parasitol. 2015;210:91–7.
Jiang J, Alderisio KA, Xiao L. Distribution of Cryptosporidium genotypes in storm event water samples from three watersheds in New York. Appl Environ Microbiol. 2005;71:4446–54.
Meerburg BG, Singleton GR, Kijlstra A. Rodent-borne diseases and their risks for public health. Crit Rev Microbiol. 2009;35:221–70.
Li W, Li Y, Song M, Lu Y, Yang J, Tao W, et al. Prevalence and genetic characteristics of Cryptosporidium, Enterocytozoon bieneusi and Giardia duodenalis in cats and dogs in Heilongjiang province, China. Vet Parasitol. 2015;208:125–34.
Zhao W, Zhang W, Yang F, Cao J, Liu H, Yang D, et al. High prevalence of Enterocytozoon bieneusi in asymptomatic pigs and assessment of zoonotic risk at the genotype level. Appl Environ Microbiol. 2014;80:3699–707.
Zhang W, Ren G, Zhao W, Yang Z, Shen Y, Sun Y, et al. Genotyping of Enterocytozoon bieneusi and subtyping of Blastocystis in cancer patients: relationship to diarrhea and assessment of zoonotic transmission. Front Microbiol. 2017;8:1835.
Yang J, Song M, Wan Q, Li Y, Lu Y, Jiang Y, et al. Enterocytozoon bieneusi genotypes in children in Northeast China and assessment of risk of zoonotic transmission. J Clin Microbiol. 2014;52:4363–7.
Xiao L, Escalante L, Yang C, Sulaiman I, Escalante AA, Montali RJ, et al. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl Environ Microbiol. 1999;65:1578–83.
Buckholt MA, Lee JH, Tzipori S. Prevalence of Enterocytozoon bieneusi in swine: an 18-month survey at a slaughterhouse in Massachusetts. Appl Environ Microbiol. 2002;68:2595–9.
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;24:4876–82.
Santín M, Fayer R. Enterocytozoon bieneusi genotype nomenclature based on the internal transcribed spacer sequence: a consensus. J Eukaryot Microbiol. 2009;56:34–8.
Feng Y. Cryptosporidium in wild placental mammals. Exp Parasitol. 2010;124:128–37.
Ziegler PE, Wade SE, Schaaf SL, Chang YF, Mohammed HO. Cryptosporidium spp. from small mammals in the New York City watershed. J Wildl Dis. 2007;43:586–96.
Chalmers RM, Robinson G, Elwin K, Hadfield SJ, Thomas E, Watkins J, et al. Detection of Cryptosporidium species and sources of contamination with Cryptosporidium hominis during a waterborne outbreak in north west Wales. J Wat Health. 2010;8:311–25.
Feng Y, Li N, Duan L, Xiao L. Cryptosporidium genotype and subtype distribution in raw wastewater in Shanghai, China: evidence for possible unique Cryptosporidium hominis transmission. J Clin Microbiol. 2009;47:153–7.
Ruecker NJ, Matsune JC, Lapen DR, Topp E, Edge TA, Neumann NF. The detection of Cryptosporidium and the resolution of mixtures of species and genotypes from water. Infect Genet Evol. 2013;15:3–9.
Fayer R, Santín M, Macarisin D. Cryptosporidium ubiquitum n. sp. in animals and humans. Vet Parasitol. 2010;172:23–32.
Blanco MA, de Lucio A, Fuentes I, Carmena D. Cryptosporidium ubiquitum in Venezuela: First report in a paediatric patient with acute diarrhoea. Enferm Infecc Microbiol Clin. 2016;34:142–3.
Langkjaer RB, Vigre H, Enemark HL, Maddox-Hyttel C. Molecular andphylogenetic characterization of Cryptosporidium and Giardia from pigs and cattle in Denmark. Parasitology. 2007;134:339–50.
Tao W, Li Y, Yang H, Song M, Lu Y, Li W. Widespread occurrence of zoonotic Cryptosporidium species and subtypes in dairy cattle from northeast China: public health concerns. J Parasitol. 2018;104:10–7.
Li P, Cai J, Cai M, Wu W, Li C, Lei M, et al. Distribution of Cryptosporidium species in Tibetan sheep and yaks in Qinghai, China. Vet Parasitol. 2016;215:58–62.
Khan SM, Debnath C, Pramanik AK, Xiao L, Nozaki T, Ganguly S. Molecular characterization and assessment of zoonotic transmission of Cryptosporidium from dairy cattle in West Bengal, India. Vet Parasitol. 2010;171:41–7.
Ong CSL, Eisler DL, Alikhani A, Fung Vicki WK, Tomblin J, Bowie WR, et al. Novel Cryptosporidium genotypes in sporadic cryptosporidiosis cases: first report of human infections with a cervinegenotype. Emerg Infect Dis. 2002;8:263–8.
Jiang Y, Tao W, Wan Q, Li Q, Yang Y, Lin Y, et al. Zoonotic and potentially host-adapted Enterocytozoon bieneusi genotypes in sheep and cattle in northeast China and an increasing concern about the zoonotic importance of previously considered ruminant-adapted genotypes. Appl Environ Microbiol. 2015;81:3326–35.
Li N, Xiao L, Wang L, Zhao S, Zhao X, Duan L, et al. Molecular surveillance of Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi by genotyping and subtyping parasites in wastewater. PLoS Negl Trop Dis. 2012;6:e1809.
Bern C, Kawai V, Vargas D, Rabke-Verani J, Williamson J, Chavez-Valdez R, et al. The epidemiology of intestinal microsporidiosis in patients with HIV/AIDS in Lima, Peru. J Infect Dis. 2015;190:1658–64.
Lobo ML, Xiao L, Antunes F, Matos O. Microsporidia as emerging pathogens and the implication for public health: a 10-year study on HIV-positive and -negative patients. Int J Parasitol. 2012;42:197–205.
Sulaiman IM, Bern C, Gilman R, Cama V, Kawai V, Vargas D, et al. A molecular biologic study of Enterocytozoon bieneusi in HIV-infected patients in Lima, Peru. J Eukaryot Microbiol. 2003;50:591–6.
Zhao W, Zhang W, Yang D, Zhang L, Wang R, Liu A. Prevalence of Enterocytozoon bieneusi and genetic diversity of ITS genotypes in sheep and goats in China. Infect Genet Evol. 2015;32:265–70.
Funding
The study was supported partially by the Graduate Student Innovation Foundation of Harbin Medical University (YJSCX2016-41HYD) and the Heilongjiang Province Education Bureau (No. 12531266). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
All data generated or analysed during this study are included in this published article. Sequences were submitted to the GenBank database under the accession numbers MG917670 and MG917671.
Author information
Authors and Affiliations
Contributions
Experiments were conceived and designed by HL and AL. Experiments were performed by WZ, J-G W, G-X R, Z-Y Y and Y-C X. The data were analyzed by WZ. FY and W-Z Z contributed reagents/materials/analysis tools. The manuscript was written by WZ, and revised by AL and HL. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
All the animals were handled and cared for according to the Chinese Laboratory Animal Administration Act of 1998. The research of protocol was reviewed and approved by the Research Ethics Committee and the Animal Ethical Committee of Harbin Medical University (HMUIRB20130009).
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Zhao, W., Wang, J., Ren, G. et al. Molecular characterizations of Cryptosporidium spp. and Enterocytozoon bieneusi in brown rats (Rattus norvegicus) from Heilongjiang Province, China. Parasites Vectors 11, 313 (2018). https://doi.org/10.1186/s13071-018-2892-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-018-2892-7