Abstract
Background
Racial/ethnic differences for commonly measured clinical variables are well documented, and it has been postulated that population-specific genetic factors may play a role. The genetic heterogeneity of admixed populations, such as African Americans, provides a unique opportunity to identify genomic regions and variants associated with the clinical variability observed for diseases and traits across populations.
Method
To begin a systematic search for these population-specific genomic regions at the phenome-wide scale, we determined the relationship between global genetic ancestry, specifically European and African ancestry, and clinical variables measured in a population of African Americans from BioVU, Vanderbilt University’s biorepository linked to de-identified electronic medical records (EMRs) as part of the Epidemiologic Architecture using Genomics and Epidemiology (EAGLE) study. Through billing (ICD-9) codes, procedure codes, labs, and clinical notes, 36 common clinical and laboratory variables were mined from the EMR, including body mass index (BMI), kidney traits, lipid levels, blood pressure, and electrocardiographic measurements. A total of 15,863 DNA samples from non-European Americans were genotyped on the Illumina Metabochip containing ~200,000 variants, of which 11,166 were from African Americans. Tests of association were performed to examine associations between global ancestry and the phenotype of interest.
Results
Increased European ancestry, and conversely decreased African ancestry, was most strongly correlated with an increase in QRS duration, consistent with previous observations that African Americans tend to have shorter a QRS duration compared with European Americans. Despite known racial/ethnic disparities in blood pressure, European and African ancestry was neither associated with diastolic nor systolic blood pressure measurements.
Conclusion
Collectively, these results suggest that this clinical population can be used to identify traits in which population differences may be due, in part, to population-specific genetics.
Similar content being viewed by others
Introduction
Racial/ethnic differences for commonly measured clinical variables, such as cholesterol [1], body mass index [2], and hypertension [3], are well documented. Although the causes of these observed differences are unclear, it has been postulated that population-specific genetic factors may play a role [4]. The genetic heterogeneity of admixed populations such as African Americans provides a unique opportunity to identify genomic regions and variants associated with the clinical variability observed for diseases and traits across populations.
Previous studies have been primarily limited to genome-wide association studies (GWAS) stratified by race/ethnicity (self-reported and/or genetic ancestry) and admixture mapping studies of one or a handful of phenotypes in mostly epidemiologic collections. Both GWAS and admixture mapping studies offer the opportunity to identify population-specific and trans-population associations involving specific genetic variants or genomic regions. Other previous studies have directly tested for associations between race/ethnicity or genetic ancestry and specific phenotypes such as atrial fibrillation [5–7]. These latter studies offer the opportunity to identify and perhaps distinguish between genetic and cultural or environmental factors that may account for the differences in disease prevalence or incidence observed across populations. Despite the success of these studies, no study has begun a systematic search of associations between genetic ancestry and traits phenome-wide.
Large epidemiologic and clinical collections often contain hundreds to thousands of data points related to the health status of individuals. To begin a systematic search for these population-specific genomic regions at the phenome-wide scale, we as the Epidemiologic Architecture for Genes Linked to Environment (EAGLE) study determined the relationship between global genetic ancestry (percent European and African ancestry) and clinical variables measured in an African American population from BioVU, the Vanderbilt University biorepository linked to de-identified electronic medical records [8, 9]. We describe here the distribution of global European and African ancestry and significantly associated clinical traits among >11,000 African Americans from BioVU. Overall, these data suggest that systematic searches for relationships between genetic ancestry and disease outcomes and traits have the potential to prioritize phenotypes with evidence of strong population differences for further study.
Methods
Study population
The DNA samples and data described here are from Vanderbilt University’s BioVU, a biorepository linked to de-identified electronic medical records. The establishment of BioVU including the ethical and legal considerations has been described elsewhere [8, 10]. Briefly, BioVU is an opt-out clinical collection that includes DNA samples extracted from discarded blood drawn for routine care at Vanderbilt University Medical Center out-patient clinics. DNA samples are linked to a de-identified version of the patient’s electronic medical records known as the Synthetic Derivative. The Synthetic Derivative contains structured, semi-structured, and unstructured clinical data that can be used for research purposes. Race/ethnicity in BioVU is administratively assigned and has been previously shown to be highly concordant with genetic ancestry for European Americans and African Americans [11, 12].
Genotyping
We as part of the EAGLE study accessed all DNA samples and data from non-European Americans within BioVU as of 2011 for genotyping. These data are collectively referred to here as “EAGLE BioVU” [9]. A total of 15,863 samples were targeted for Illumina Metabochip genotyping. The Illumina Metabochip is a 200,000 variant array designed for replicating genome-wide association study findings (index variants) and for fine mapping select GWAS findings for cardiovascular and metabolic traits and outcomes [13]. The EAGLE BioVU dataset was generated by the Vanderbilt DNA Resources Core, and genotype calls and quality control were performed by the Population Architecture using Genomic and Epidemiology (PAGE) Coordinating Center as previously described [9, 14].
Phenotyping
We defined 36 phenotypes using a combination of billing (International Classification of Diseases 9 or ICD-9) codes, procedure (CPT) codes, labs, and clinical notes available in the Synthetic Derivative. All phenotypes described here were extracted for the genetic association studies as part of the larger PAGE I study [15] and will be available via dbGaP. Algorithms for body mass index [Goodloe R, Faber-Eger E, Boston J, Crawford DC, Bush WS: Reducing clinical noise for body mass index measures due to unit and transcription errors in the electronic medical record, in preparation]. electrocardiographic traits (QRS duration, PR interval, QT interval, QRS, and heart rate) [16], and type 2 diabetes [17] have been previously described. The other phenotypes were defined as follows:
Lipids
-
Laboratory measurements were queried for high-density lipoprotein cholesterol (HDL-C), low density lipoprotein cholesterol (LDL-C), total cholesterol, and triglycerides.
-
Records were also queried for calculated LDL-C.
-
For each individual, median values were calculated for a) measurements taken when no medications are prescribed (“pre-medication” values) and b) measurements taken at first mention of medication and post mention of medication (“post-medication” values)
-
Medication class and list: statins (also known as HMG CoA reductase inhibitors, atorvastatin (Lipitor®), fluvastatin (Lescol®), lovastatin (Mevacor®, Altoprev™), pravastatin (Pravachol®), rosuvastatin calcium (Crestor®), simvastatin (Zocor®), lovastatin + niacin (Advicor®), atorvastatin + amlodipine (Caduet®), and simvastatin + ezetimibe (Vytorin™); selective cholesterol absorption inhibitors (ezetimibe (Zetia®)); resins (cholestyramine (Questran®, Questran® Light, Prevalite®, Locholest®, Locholest® Light), colestipol (Colestid®), colesevelam Hcl (WelChol®)); fibrates (gemfibrozil (Lopid®), fenofibrate (Antara®, Lofibra®, Tricor®, and Triglide™), clofibrate (Atromid-S)); and niacin.
Kidney traits
-
Laboratory measures were queried for albumin (UABM), serum albumin (ALB), urinary albumin (UAlb, AlbCnc), albumin/creatinine ratio (AlbCre, SUA/C), creatinine from blood (Creat), creatinine from urine (URCRE, SUCrea, Creat1), urea nitrogen blood (BUN), and uric acid.
-
For each individual, median values were calculated if more than one value was available in the Synthetic Derviative.
Primary essential hypertension
-
Case definition 1: Individuals on hypertensive medication (list under systolic and diastolic blood pressure) and ICD-9 codes 401.* or mention of “high blood pressure” or “hypertension” in problem list
-
Case definition 2: individuals not on hypertensive medication (list under systolic and diastolic blood pressure) and >140/90 systolic/diastolic blood pressure readings
-
“Baseline” hypertension was defined as an individual’s hypertension status at first clinical visit.
-
“Lifetime” hypertension status was defined as an individual’s hypertension status over the course of the clinical records available in the Synthetic Derivative.
Systolic and diastolic blood pressure
-
Measurements for pregnant individuals were excluded.
-
In-patient measurements were excluded.
-
For each individual, “baseline” blood pressure measures represent the first systolic and diastolic blood pressure mentioned in the Synthetic Derivative.
-
For each individual, median values were calculated for a) measurements taken when no medications are prescribed (“pre-medication” values) and b) measurements taken at first mention of medication and post mention of medication (“post-medication” values).
-
Medications: angiotensin converting enzyme inhibitors, angiotensin receptor blockers, beta blockers, non-dihydropyridine calcium channel blockers, dihydropyridine calcium channel blockers, hydralazine, minoxidil, central alpha agonists, direct renin antagonists, aldosterone antagonists, alpha antagonists, diuretics (thiazides, K-sparing, and loop diuretics)
-
Excluded medication: phentolamine, phenoxybenzamine
Type 2 diabetes related traits
-
Laboratory measures were queried for glucose, glycated hemoglobin, and insulin.
-
For each individual, median values were calculated if more than one value was available in the Synthetic Derivative.
Statistical methods
EAGLE BioVU Metabochip genotype data as part of the PAGE I study were first subjected to quality control, including the identification of genetic ancestry outliers via EIGENSTRAT, through the PAGE I Coordinating Center [14]. We selected 7,655 uncorrelated SNPs from among the ancestry informative SNPs described by the PAGE I Coordinating Center [14] assayed by the Illumina Metabochip to estimate global genetic ancestry among administratively assigned African Americans in quality-controlled EAGLE BioVU genotype data using STRUCTURE v2.3.4 (K = 3) [18]. Data from 395 International HapMap samples represented CEU, YRI, and CHB/JPN were downloaded from the International HapMap Project and included in STRUCTURE runs as predefined parental clusters. Percent European ancestry was determined by STRUCTURE for each individual and used as the independent variable in tests of association.
Tests of association were performed using logistic or linear regression for 36 outcomes or traits where global ancestry was the independent variable. Primary essential hypertension (baseline and lifetime) and type 2 diabetes were binary outcomes; all other outcomes were continuous. Similar to the high-throughput PheWAS pipeline proposed by the PAGE I study [19], analyses were not adjusted for covariates. Also similar to the PAGE I study PheWAS pipeline [19], all continuous traits were tested for an association as untransformed and transformed (1+ natural logarithm) variables. Analyses were performed using SAS version 9.2 (Cary, NC) and R (version 3.2.0) in R Studio (version 0.99.441).
We estimated local ancestry using LAMP [20] for three fine-mapped regions on the Metabochip representing the three strongest associations with global European ancestry ranked by p-values from the tests of association: QRS duration, QT interval, and BMI. For these three traits, we identified a GWAS-significant variant in the NHGRI GWAS Catalog [21] that was assayed by the Metabochip and further fine-mapped by the Metabochip. From this search, we identified SCN10A rs6801957 (QRS duration) [22], NOS1AP rs12143842 (QT interval) [23–27], and FTO rs1558902 (BMI) [28]. We then considered assayed genetic variants within a 50 kb window of each of these GWAS-index variants (SCN10A chr3:38664112–38860101; NOS1AP chr1:160256929–160654852; FTO chr16:52245615–52755879) for local ancestry estimation using LAMP assuming 10 generations with an alpha at 0.2 and 0.8 and with a recombination rate of 1.0x10−9. We tested for associations between percent local European and African ancestry and each of the three traits untransformed and transformed (1+ natural logarithm).
Results
EAGLE BioVU characteristics are given in Table 1. On average, most are female and relatively young. The largest non-European descent group is African American (n = 11,166), consistent with Davidson County, Tennessee population characteristics based on the 2010 US Census. The average individual in EAGLE BioVU had approximately 82 clinic visits and 147 ICD-9 codes available in his/her medical record.
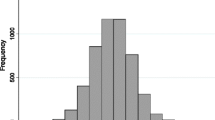
For each African American in EAGLE BioVU, we estimated percent European global genetic ancestry using 7,655 SNPs from the Metabochip and STRUCTURE. As expected, the African American population in EAGLE BioVU represents a complex, two-way admixture event of European and African-descent populations (Figs. 1, 2 and 3). Percent West African ancestry ranged from 0 % to 100 % with a mean of 81 % and interquartile range of 12.7 % (Fig. 3). Percent European ancestry ranged from 0 % to 100 % with a mean of 17.2 % and interquartile range of 12.8 %. The average percent European ancestry estimated here is consistent with other estimates for African Americans [5, 7].
Genetic ancestry and EAGLE BioVU African Americans. Global genetic ancestry was estimated in 11,166 African Americans in EAGLE BioVU using 7,655 SNPs from the Illumina Metabochip and STRUCTURE assuming K = 3. The STRUCTURE plot is anchored by 395 International HapMap Project samples (CEU in red, YRI in green, and CHB/JPT in light blue)
Distribution of percent European global ancestry among. African Americans in EAGLE BioVU. STRUCTURE was used to estimate global genetic ancestry using 7,655 SNPs assayed on the Illumina Metabochip and assuming K = 3. Plotted are the global European ancestry estimates for 11,166 African Americans in EAGLE BioVU where the x-axis represents the % global European ancestry and the y-axis represents the % of the total African American sample in EAGLE BioVU
Distribution of percent African global ancestry among. African Americans in EAGLE BioVU. STRUCTURE was used to estimate global genetic ancestry using 7,655 SNPs assayed on the Illumina Metabochip and assuming K = 3. Plotted are the global West African ancestry estimates for 11,166 African Americans in EAGLE BioVU where the x-axis represents the % global African ancestry and the y-axis represents the % of the total African American sample in EAGLE BioVU
Given that race/ethnicity is administratively assigned and not self-reported [11, 12], it is possible that individuals at the extremes of the global genetic ancestry distribution have been incorrectly assigned race/ethnicity. To explore this possibility, we extracted country of origin data from the clinical notes of EAGLE BioVU [Farber-Eger E, Goodloe R, Boston J, Bush WS, Crawford DC: Extracting country-of-origin from electronic medical records for gene-environment studies as part of the Epidemiologic Architecture for Genes Linked to Environment (EAGLE) study, in preparation]. Of the 1,159 individuals with less than 5 % European global genetic ancestry, only 44 had data on country of origin. Of these 44, 25 individuals with less than 5 % European global genetic ancestry have clinical notes with evidence that they are from African nations (Burundi, Ethiopia, Ghana, Kenya, Malawi, Nigeria, Rwanda, Senegal, Somalia, Sudan, Tanzania, and Uganda). Of the 114 individuals with >95 % European global genetic ancestry, only one individual had country of origin data (Haiti).
We then performed sex-combined tests of association using logistic or linear regression for 36 outcomes or traits where European global ancestry was the independent variable (Table 2). Among the three binary outcomes considered, percent European ancestry was not associated with baseline hypertension (56 %; p = 0.21), lifetime hypertension (78 %; p = 0.39), or type 2 diabetes (12 %; p = 0.18). Among the remaining continuous outcomes considered, only QRS duration was significantly associated with percent European global genetic ancestry (p = 6.7x10-5, n = 837; Fig. 4). European ancestry was not associated with blood pressure measurements, kidney traits, type 2 diabetes associated measures, or any of the lipid traits (Table 2; Fig. 4). When all continuous traits were transformed, similar results were observed where only QRS duration was significantly associated with European global ancestry (p = 9.15x10−5; Additional file 1: Table S1).
Continuous outcomes tested for an association with global European ancestry in African Americans from EAGLE BioVU. Tests of association were performed using linear regression between 33 continuous outcomes and percent. European global genetic ancestry among African Americans in EAGLE BioVU. Plotted are the –log10 of the p-value and the effect size (beta). Significant results are annotated in blue whereas non-significant results are annotated in gray. The red line represents the significance threshold at p = 0.001
We also performed sex-combined tests of association using logistic or linear regression for the same 36 outcomes or traits where African global ancestry was the independent variable (Table 3; Additional file 1: Table S2). Like the European global ancestry analyses (Table 2), the only significant association identified was for QRS duration (p = 1.3x10−4; Fig. 5). The genetic effect for the association identified between African global ancestry and QRS duration (β = −7.45; standard error 1.93) was similar in magnitude but opposite in direction compared with the association identified for European global ancestry (β = 7.98; standard error = 1.99; Table 2; Fig. 4).
Continuous outcomes tested for an association with global African ancestry in African Americans from EAGLE BioVU. Tests of association were performed using linear regression between 33 continuous outcomes and percent. African global genetic ancestry among African Americans in EAGLE BioVU. Plotted are the –log10 of the p-value and the effect size (beta). Significant results are annotated in blue whereas non-significant results are annotated in gray. The red line represents the significance threshold at p = 0.001
For the three most significant test of associations between global ancestry and the outcomes and traits considered here (QRS duration, QT interval, and BMI; Table 2), we estimated local ancestry using LAMP [20] in the three fine-mapped regions (SCN10A, NOS1AP, and FTO) and tested for associations with local European and African ancestry. In this sample of African Americans, the QT interval associated NOS1AP region had on average the least European ancestry (16.7 %) compared with the BMI associated FTO region (23.6 %) and the QRS duration associated SCN10A region (24.6 %). None of the tests of association between local ancestry and the three traits was significant at p < 0.05.
Discussion
We extracted 36 health outcomes and traits from de-identified electronic medical records of 11,166 African Americans in EAGLE BioVU and tested each of these phenotypes for an association with percent European and percent African global genetic ancestry. In sex-combined analyses, we identified a significant association between QRS duration and both European and African ancestry albeit in opposite directions. Other than QRS duration, no other outcome or trait was associated with European or African ancestry in sex-combined analyses.
QRS duration is a cardiac conduction trait extracted from electrocardiograms. Normal QRS duration generally ranges from 70 to 100 milliseconds in most populations [16]. Abnormal or prolonged QRS duration (>120 msec) is used in the diagnosis of bundle branch block or ventricular rhythm. The significant association identified here with European and African ancestry is consistent with previous observations that African Americans tend to have a shorter QRS duration compared with European Americans [16]. The lack of association between global ancestry and QT interval, another cardiac conduction trait tested here, is consistent with a similar study in African Americans from seven large population-based cohorts [25].
The present study had several limitations and strengths. A major limitation of this study is that global ancestry as opposed to local ancestry was estimated for each individual. The estimation of local ancestry will enable the identification of associations between phenotypes and specific genomic regions. However, estimation of local ancestry requires computational resources and dense genome-wide data. This dataset was limited to Metabochip data, which is dense only in fine-mapped regions and is sparse in non-fine mapped regions of the genome. Given the uneven genome-wide coverage, we were unable to confidently impute genome-wide data nor estimate local genetic ancestry at a genome-wide level for further study.
Despite these limitations, we did estimate local ancestry for three specific regions associated with three specific traits including QRS duration. In this sample of African Americans, QRS duration was not associated with local ancestry estimates for the SCN10A fine-mapped region. Variants in SCN10A have been associated with cardiac conduction (PR interval, QRS duration, and QT interval) in multiple populations including European-descent [22, 29–31] and Indian Asians [29]. For African Americans, variants in SCN10A have been strongly associated with PR interval [32]. Although a small GWAS in African Americans did not identify genome-wide significant associations between QRS duration and variants in SCN10A, the data suggest that the GWAS-index variants identified in European and Indian Asian populations generalize to African Americans with a similar genetic effect size and direction [33]. The lack of association between local genetic ancestry at SCN10A and QRS duration may be due to the fact that this locus is not responsible for the global ancestry signal detected here. This negative finding coupled with the lack of powerful GWAS in African Americans for QRS duration suggest that there may be other genetic variants and gene regions yet to be associated with this cardiac conduction trait in this population.
Another potential limitation for this global ancestry PheWAS is sample size and power. Overall, the sample size is moderate, with >11,000 African Americans with health-related data available for study. For individual phenotypes, however, sample size and power vary. The most powerful tests of association were limited to the laboratory values or vital signs routinely collected by the clinic, such as total cholesterol and blood pressure. However, even for these common measures, power may be adversely impacted by imprecise phenotype efforts when extracting data from electronic medical records. For example, it is assumed but not known if laboratory measures such as the lipid traits or type 2 diabetes traits were taken while the individual was fasting. Likewise, blood pressure measurements can be impacted by prescription medication noncompliance and diurnal effects. While care is taken to account for these factors in the phenotyping process, it is likely that all phenotypes extracted from electronic medical records have some degree of unintended phenotypic heterogeneity that will impact statistical power.
A major strength of this study is that it is one of the few large, clinical collections available for admixed populations such as African Americans. BioVU continues to accrue clinical data linked to DNA samples already collected as well as clinical data for new DNA samples representing new patients entering the Vanderbilt University Medical Center system. The accrual of these new data, samples, and eventually genetic data make this a potentially powerful dataset for further research in genetics and health disparities. Another major strength of the current study is electronic phenotyping using structured and unstructured data available in the EMR. Most previous PheWAS in clinical populations with available EMR data have been limited to billing (ICD-9) codes [30, 34–36]. Here, we take full advantage of the richness of the EMR to define cases and controls for select binary traits as well as to extract laboratory values for consideration as outcome variables. The specific variables for phenotyping and analysis in this PheWAS were chosen based on anticipated downstream studies in PAGE I [15]. While 36 phenotypes represent only a fraction of the phenome compared with a PheWAS based on ICD-9 codes, many of the phenotypes considered here have not yet been explored in published PheWAS. Also, limiting this PheWAS to 36 phenotypes reduces the number of statistical tests performed, which in turn lessens the impact of multiple testing when interpreting statistical significance of the results. The association observed between European global ancestry and QRS duration at p = 6.7x10−5 survives correction for multiple testing even if a conservative Bonferroni correction (p = 0.0014) is applied.
Conclusions
We estimated global genetic ancestry in an admixed population and systematically searched for associations between European and African ancestry and clinical outcomes and traits mined from electronic medical records. We identified a significant association between European and African ancestry and QRS duration, and this association is supported by the known epidemiology of this electrocardiographic trait in diverse populations. Further general genomic discovery and study of local genetic ancestry are warranted to identify specific genomic regions associated with clinical outcomes in diverse populations.
References
Carroll MD, Lacher DA, Sorlie PD. Trends in serum lipids and lipoproteins of adults, 1960–2002. JAMA. 2005;294:1773–81.
Flegal KM, Carroll MD, Kit BK, Ogden CL. Prevalence of obesity and trends in the distribution of body mass index among us adults, 1999–2010. JAMA. 2012;307:491–7.
Egan BM, Zhao Y, Axon R. US trends in prevalence, awareness, treatment, and control of hypertension, 1988–2008. JAMA. 2010;303:2043–50.
Burchard EG, Ziv E, Coyle N, Gomez SL, Tang H, Karter AJ, et al. The importance of race and ethnic background in biomedical research and clinical practice. N Engl J Med. 2003;348:1170–5.
Marcus GM, Alonso A, Peralta CA, Lettre G, Vittinghoff E, Lubitz SA, et al. European ancestry as a risk factor for atrial fibrillation in African Americans. Circulation. 2010;122:2009–15.
Delaney JT, Jeff JM, Brown NJ, Pretorius M, Okafor HE, Darbar D, et al. Characterization of genome-wide association-identified variants for atrial fibrillation in African Americans. PLoS ONE. 2012;7:e32338.
Perez MV, Hoffmann TJ, Tang H, Thornton T, Stefanick ML, Larson JC, et al. African American race but not genome-wide ancestry is negatively associated with atrial fibrillation among postmenopausal women in the Women’s Health Initiative. Am Heart J. 2013;166:566–72.
Roden DM, Pulley JM, Basford MA, Bernard GR, Clayton EW, Balser JR, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther. 2008;84:362–9.
Crawford DC, Goodloe R, Farber-Eger E, Boston J, Pendergrass SA, Haines JL, et al. Leveraging epidemiologic and clinical collections for genomic studies of complex traits. Hum Hered. 2015;79:137–46.
Pulley J, Clayton E, Bernard GR, Roden DM, Masys DR. Principles of human subjects protections applied in an opt-out, de-identified biobank. Clin Transl Sci. 2010;3:42–8.
Dumitrescu L, Ritchie MD, Brown-Gentry K, Pulley JM, Basford M, Denny JC, et al. Assessing the accuracy of observer-reported ancestry in a biorepository linked to electronic medical records. Genet Med. 2010;12:648–50.
Hall JB, Dumitrescu L, Dilks HH, Crawford DC, Bush WS. Accuracy of administratively-assigned ancestry for diverse populations in an electronic medical record-linked biobank. PLoS ONE. 2014;9:e99161.
Voight BF, Kang HM, Ding J, Palmer CD, Sidore C, Chines PS, et al. The Metabochip, a custom genotyping array for genetic studies of metabolic, cardiovascular, and anthropometric traits. PLoS Genet. 2012;8:e1002793.
Buyske S, Wu Y, Carty CL, Cheng I, Assimes TL, Dumitrescu L, et al. Evaluation of the Metabochip genotyping array in African Americans and implications for fine mapping of GWAS-identified loci: the PAGE study. PLoS ONE. 2012;7:e35651.
Matise TC, Ambite JL, Buyske S, Carlson CS, Cole SA, Crawford DC, et al. The next PAGE in understanding complex traits: design for the analysis of Population Architecture Using Genetics and Epidemiology (PAGE) Study. Am J Epidemiol. 2011;174:849–59.
Ramirez AH, Schildcrout JS, Blakemore DL, Masys DR, Pulley JM, Basford MA, et al. Modulators of normal electrocardiographic intervals identified in a large electronic medical record. Heart Rhythm. 2011;8:271–7.
Kho AN, Hayes MG, Rasmussen-Torvik L, Pacheco JA, Thompson WK, Armstrong LL, et al. Use of diverse electronic medical record systems to identify genetic risk for type 2 diabetes within a genome-wide association study. J Am Med Inform Assoc. 2012;19:212–8.
Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–59.
Pendergrass SA, Brown-Gentry K, Dudek SM, Torstenson ES, Ambite JL, Avery CL, et al. The use of phenome-wide association studies (PheWAS) for exploration of novel genotype-phenotype relationships and pleiotropy discovery. Genet Epidemiol. 2011;35:410–22.
Pasaniuc B, Sankararaman S, Kimmel G, Halperin E. Inference of locus-specific ancestry in closely related populations. Bioinformatics. 2009;25:i213–21.
Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–6.
Sotoodehnia N, Isaacs A, de Bakker PIW, Dorr M, Newton-Cheh C, Nolte IM, et al. Common variants in 22 loci are associated with QRS duration and cardiac ventricular conduction. Nat Genet. 2010;42:1068–76.
Newton-Cheh C, Eijgelsheim M, Rice KM, de Bakker PIW, Yin X, Estrada K, et al. Common variants at ten loci influence QT interval duration in the QTGEN Study. Nat Genet. 2009;41:399–406.
Pfeufer A, Sanna S, Arking DE, Muller M, Gateva V, Fuchsberger C, et al. Common variants at ten loci modulate the QT interval duration in the QTSCD Study. Nat Genet. 2009;41:407–14.
Smith JG, Avery CL, Evans DS, Nalls MA, Meng YA, Smith EN, et al. Impact of ancestry and common genetic variants on QT interval in African Americans. Circ Cardiovasc Genet. 2012;5:647–55.
Avery CL, Sethupathy P, Buyske S, He Q, Lin DY, Arking DE, et al. Fine-mapping and initial characterization of QT interval loci in African Americans. PLoS Genet. 2012;8:e1002870.
Seyerle AA, Young AM, Jeff JM, Melton PE, Jorgensen NW, Lin Y, et al. Evidence of heterogeneity by race/ethnicity in genetic determinants of QT interval. Epidemiology. 2014;25:790–8.
Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–48.
Chambers JC, Zhao J, Terracciano CMN, Bezzina CR, Zhang W, Kaba R, et al. Genetic variation in SCN10A influences cardiac conduction. Nat Genet. 2010;42:149–52.
Ritchie MD, Denny JC, Zuvich RL, Crawford DC, Schildcrout JS, Bastarache L, et al. Genome- and phenome-wide analyses of cardiac conduction identifies markers of arrhythmia risk. Circulation. 2013;127:1377–85.
Verweij N, Leach IM, van den Boogaard M, van Veldhuisen DJ, Christoffels VM, LifeLines CS, et al. Genetic determinants of P wave duration and PR segment. Circ Cardiovasc Genet. 2014;7:475–81.
Smith JG, Magnani JW, Palmer C, Meng YA, Soliman EZ, Musani SK, et al. Genome-wide association studies of the PR interval in African Americans. PLoS Genet. 2011;7:e1001304.
Jeff JM, Ritchie MD, Denny JC, Kho AN, Ramirez AH, Crosslin D, et al. Generalization of variants identified by genome-wide association studies for electrocardiographic traits in African Americans. Ann Hum Genet. 2013;77:321–32.
Denny JC, Ritchie MD, Basford MA, Pulley JM, Bastarache L, Brown-Gentry K, et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics. 2010;26:1205–10.
Denny JC, Bastarache L, Ritchie MD, Carroll RJ, Zink R, Mosley JD, et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotech. 2013;31:1102–11.
Cronin RM, Field JR, Bradford Y, Shaffer CM, Carroll RJ, Mosley JD, et al. Phenome wide association studies demonstrating pleiotropy of genetic variants within FTO with and without adjustment for body mass index. Front Genet. 2014;5:250.
Acknowledgments
This work was supported in part by NIH U01 HG004798 and its ARRA supplements. The dataset(s) used for the analyses described were obtained from Vanderbilt University Medical Center’s BioVU which is supported by institutional funding and by the Vanderbilt CTSA grant funded by the National Center for Research Resources, Grant UL1 RR024975-01, which is now at the National Center for Advancing Translational Sciences, Grant 2 UL1 TR000445-06.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
LD, WSB, SAP, and DCC designed the study. RG, JB, and EF-E extracted the data. LD and NAR performed the analyses. LD and DCC drafted the manuscript. DCC obtained funding for the project. All authors read and approved the final manuscript.
Additional file
Additional file 1: Table S1.
Transformed continuous outcomes tested for an association with global European ancestry in African Americans from EAGLE BioVU. Tests of association were performed using linear regression between 33 transformed (1 + natural logarithm) continuous outcomes and percent European global genetic ancestry among African Americans in EAGLE BioVU (n = 11,166). Test results of all associations are shown for each outcome tested. Significant associations are bolded and italicized. Abbreviations: odds ratio (OR), standard deviation (SD), and standard error (SE). Table S2. Transformed continuous outcomes tested for an association with global African ancestry in African Americans from EAGLE BioVU. Tests of association were performed using linear regression between 33 transformed (1 + natural logarithm) continuous outcomes and percent African global genetic ancestry among African Americans in EAGLE BioVU (n = 11,166). Test results of all associations are shown for each outcome test. Significant associations are bolded and italicized. Abbreviations: odds ratio (OR), standard deviation (SD), and standard error (SE). (DOCX 23 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Dumitrescu, L., Restrepo, N.A., Goodloe, R. et al. Towards a phenome-wide catalog of human clinical traits impacted by genetic ancestry. BioData Mining 8, 35 (2015). https://doi.org/10.1186/s13040-015-0068-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13040-015-0068-y