Abstract
Background
In view of previous conflicting findings, this meta-analysis was performed to comprehensively determine the overall strength of associations between brain-derived neurotrophic factor (BDNF) genetic polymorphism Val66Met and susceptibility to bipolar disorders (BPD).
Methods
Literatures published and cited in Pubmed and Wanfang Data was searched with terms of ‘Val66Met’, ‘G196A’, ‘rs6265’, ‘BDNF’, ‘association’, and ‘bipolar disorder’ up to March 2014. All original case–control association studies were meta-analyzed with a pooled OR to estimate the risk and 95% confidence interval (CI) to reflect the magnitude of variance.
Results
Twenty-one case–control association studies met our criteria for the meta-analysis. Overall, there was no significant difference in allelic distribution of Val66Met polymorphism between patients and controls with a pooled OR = 1.03 (95% CI 0.98, 1.08) although there was a trend towards association between Val66Met polymorphism and BPD in Caucasians with an OR of 1.08 (95% CI 1.00, 1.16). However, subgroup analyses showed that there was a significant association of Val allele with decreased disease susceptibility for bipolar disorder type II with a pooled OR of 0.88 (95% CI 0.78, 0.99).
Conclusions
There is no compelling evidence to supportVal66Met polymorphism in BDNF gene playing an important role in the susceptibility to BPD across different ethnicities.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
Bipolar disorders (BPD) are chronic, recurrent, debilitating disorders with high lifetime prevalence and significant disease burden across different populations [1]-[3]. However, recent advances in pharmacological treatment for BPD remained quite modest. The treatment of bipolar depression is still a major challenge [4],[5]. Moreover, BPD is frequently unrecognized and misdiagnosed, particularly in patients presenting with their first-episode of depression. These patients are often treated with inappropriate and costly regimens [6]-[9]. Thus, there is an urgent need to understand the pathophysiology of BPD in order to develop earlier diagnoses and more effective treatments [10]. Family, twins and epidemiological studies unequivocally demonstrate that BPD is a highly heritable disease with a heritability of more than 85%, and involves the interaction of multiple genes or more complex genetic mechanisms [11]-[13]. To date, association studies support a possible role for several candidate genes in BPD, including brain-derived neurotrophic factor (BDNF), but consistent direction of effects and alleles have not been established [14]. These inconsistent findings from previous genetic association studies may be related to variation in ascertainment, phenotype definition and control selection, limited power and possibly confounded by ethnic heterogeneity and population substructure [10],[15].
The hypothesis of neuronal plasticity involved in mood disorders has been supported by the use of antidepressants and mood stabilizers, e.g. lithium and valproate, inducing the expression of neurotrophins (e.g. BDNF) and synaptic changes [16],[17]. Moreover, BDNF gene has been implicated in the etiology of BPD by linkage studies [18]. Position 196 in exon 5 of the BDNF gene contains a G to A transition (dbSNP: rs6265) that results in an amino acid substitution (valine to methionine) at codon 66 in the precursor BDNF peptide sequence [19]. This change results in BDNF functional polymorphisms in this region as Val66Val, Val66Met, and Met66Met. Two previous family-based association studies found that this functional polymorphism Val66Met was significantly associated with the susceptibility to BPD [20],[21]. Following these two reports, a large number of association studies between BDNF gene polymorphisms and BPD have been published. Most of them specifically focused on the Val66Met polymorphism, but yielded conflicting results [10],[15],[22]. In view of the conflicting results, a meta-analysis on all original case–control association studies was performed to comprehensively determine the overall strength of associations between BPD and Val66Met polymorphism.
Methods
Literature search
Studies included in the analysis were searched from two databases: Pubmed (http://www.ncbi.nlm.nih.gov/pubmed/) and Wanfang Data (http://www.wanfangdata.com/), with the keywords ‘Val66Met’, ‘G196A’, ‘rs6265’, ‘BDNF’, ‘association’, and ‘bipolar disorder’ in varying combinations. The retrieved abstracts were used to identify studies that examined the allelic association between the Val66Met polymorphism of BDNF and bipolar disorder. Bibliographies or citations from retrieved articles were also cross-referenced as well. The searched period was from the first data available in each database up to March 2014. Two independent authors extracted the following data from each eligible study: last name of the first author, year of publication, ethnicity, sample sizes and allele frequencies of cases and controls, etc. Discrepancies were resolved by mutual consent.
All eligible studies were determined against the following inclusion criteria: (i) published in a peer-reviewed journal; (ii) presented original data; (iii) provided either allele frequency of Val(G)/Met(A), or genotypes (Val/Val, Val/Met, Met/ Met) in both BPD patients and healthy controls; (iv) enrolled more than 100 subjects in both patients group and controls group; and (v) designed as a case–control study. Both family-based studies and genome-wide association studies were excluded in this research. Duplications were deleted, as well as studies that reported all or part of their data previously. The authors of studies were contacted for additional information (e.g. allele or genotype frequencies or characteristics of the samples) if there was uncertainty about whether their data met our inclusion–exclusion criteria, or if we needed additional data which were not contained in the original report.
Statistical analyses
Data were classified by diagnostic category (case or control) and allele (Val or Met), and Val was assigned as the risk allele. Meta-analysis was performed similar to that described previously [22]. The pooled OR was calculated according to the methods of DerSimonian [23], and its 95% confidence interval (CI) was constructed using Woolf’s method [24]. The Cochran chi-square-based Q statistical test was performed to assess the heterogeneity of ORs, and the significance of the pooled OR was determined by the z-test. If the result of the heterogeneity test was p ≥ 0.05, ORs were pooled according to the fixed-effects model (Mantel-Haenszel methods); otherwise, the random-effects model was used. All statistical analyses were conducted using Review Manager Version 5.2 (RevMan 5.2) [25]. A sensitivity analysis of one-study removed strategy was used to evaluate whether or not the results are being driven by any one specific study, and a funnel plot was used to detect whether or not there is evidence of publication bias. Statistical tests were two-tailed, and the significance level was set at P < 0.05, unless stated otherwise.
Results
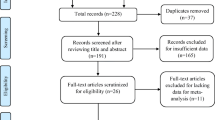
The process of identifying studied included in this meta-analysis is shown in Figure 1. Twenty-one case–control association studies met our criteria for the meta-analysis (Table 1). Data from four studies [26]-[29] were excluded due to the partial overlap with a larger sample size case–control study [30], and data from six studies were excluded due to less than 100 subjects in either patient group or control group [31]-[36]. Additionally, the case–control sample from a genome-wide association study was also excluded from current meta-analysis [37].
Overall, the data from 7219 BPD cases and 9832 healthy controls were analyzed. The mean genotype distribution in Caucasian and Oriental population was presented in Table 2. There was no significant difference in allelic distribution of Val66Met polymorphism between patients and controls. The pooled OR was 1.03 (95% CI: 0.98-1.08, Z = 1.00, P = 0.32) (Figure 2). Similarly, there was also no significant difference in allelic distribution of Val66Met polymorphism between patients and controls in Oriental population, with a pooled OR of 0.96 (95% CI: 0.89-1.05, Z = 0.82, P = 0.41) for Han Chinese population, 0.99 (95% CI: 0.85-1.15, Z = 0.12, P = 0.90) for Japanese population, and 1.06 (95% CI: 0.89-1.25, Z = 0.67, P = 0.50) for Korean population, respectively (Figure 2). However, there was a trend towards significant difference in Caucasian population with a pooled OR of 1.08 (95% CI: 1.00-1.16, Z = 1.97, P = 0.05) (Figure 2). The sensitivity analysis showed that the results were not being driven by any one specific study, and the funnel plot did not detect there was evidence of publication bias (Figure 3).
Among 21 included studies, six case–control association studies made a distinction for clinical phenotypes between bipolar I disorder (BP I) and bipolar II disorder (BP II), and two other studies only recruited BP I patients (Table 3). A further meta-analysis of the data from the aforementioned eight studies did not find a significant difference in allelic distribution of Val66Met polymorphism between BP I patients and healthy controls with a pooled OR of 1.00 (95% CI: 0.93-1.08, Z = 0.04, P = 0.97) (Figure 4). However, there was a significant difference in allelic distribution of Val66Met polymorphism between BP II patients and healthy controls with a pooled OR of 0.88 (95% CI: 0.78-0.99, Z = 2.20, P = 0.03) (Figure 4). A post-hoc analysis did not find a significant difference in allelic distribution of Val66Met polymorphism between BP I patients and BP II patients with a pooled OR of 1.10 (95% CI: 0.98-1.25, Z = 1.58, P = 0.12) (Figure 5).
Discussion
With a total of 7,219 patients and 9,832 control cases, our meta-analysis included an additional 4,076 BPD cases and 3,485 healthy controls compared to a previous meta-analysis of case–control studies [22]. Similar to this previous meta-analysis, we did not find significant associations between the Val66Met polymorphism and BPD susceptibility either in a combined population or a subgroup of Oriental patients. However, there was a trend towards significant association in Caucasian population (fixed-effects pooled OR = 1.08, P = 0.05). Subgroup meta-analyses also showed that the Val allele may be a protective factor for BP II (fixed-effects pooled OR = 0.88, P = 0.03).
Our overall finding, that no convincing evidence for association between the Val66Met polymorphism and BPD as a whole, is consistent with a previous meta-analysis reported by Kanazawa et al. [22], which included 11 case–control designed studies up to February 2006. However, our overall finding is inconsistent with the result of a meta-analysis conducted by Fan et al. [15], which included all original case–control and family-based association studies published up to May 2007 and found a modest but statistically significant association between the Val allele and BPD susceptibility [15]. Both our analysis and the analysis of Kanazawa et al. [22] exclusively included case–control studies. In contrast, the analysis of Fan et al. [15] included five family-based association studies with a total of 858 families and a genome-wide association study with 1866 BPD patients and 2932 controls. The inclusion of different studies in these meta-analyses may explain the discrepancy among these three meta-analysis studies. It is well known that BPD is a highly inheritable disorder. Inclusion of family-based association studies in the meta-analysis of Fan et al. [15] might increase the probability of detecting difference between patients and controls.
The finding of insignificant association between the Val66Met polymorphism and BPD in Oriental population populations (Chinese, Japanese and Korean populations), but a trend towards significance in Caucasian population suggest that ethnic heterogeneity may affect the results of these genetic association studies. Fan et al. [15] reported that the allele frequencies of the Val66Met polymorphism in BDNF gene across individual studies and four HapMap populations (European, Chinese, Japanese and Yoruban populations) had significant global variations, which raised concerns of possible population stratification among case–control studies [15]. A more recent population genetic study found that there were substantial variations in BDNF coding regions and haplotype frequencies between 58 global populations with the Met allele of Val66Met ranged from 0-72% frequencies [58]. As previously pointed out, unless all study participants are from a homogenous ethnic group, the confounding effect from different ethnic groups is inevitable [10]. Moreover, control selection (e.g. healthy control or family control) might also result in the discrepancy of findings in these genetic association studies. A significant discrepancy between the pooled ORs from case–control studies and family-based studies raised a concern regarding a more generalized transmission distortion at this locus that is not disease related [15].
The finding of significant associations between Val66Met polymorphism and BP II suggests that the strength of association between Val66Met polymorphism and BPD could depend on the clinical phenotypes or subtypes of BPD such as with rapid cycling course, early onset, or substance comorbidity [10],[28],[30]-[32],[34],[50],[55],[59]. Interestingly, the Val allele may have opposite associations with disease susceptibility in different bipolar subtypes. The findings of two previous studies in Caucasian population (a case–control study and a family-based study) appeared that the Val allele was associated with an increased disease risk for rapid-cycling bipolar disorder (RCBD) [46],[60], but this meta-analysis showed a decreased risk for BP II (especially in Han Chinese population). However, another family-based association analysis in Caucasian population did not replicate a significant association between the Val66Met polymorphism and BPD or RCBD. Thus the discrepancy of findings in these studies may stem from the differences of clinical phenotypes, ethnic origin and control selection.
Several limitations of this meta-analysis should be considered. One important limitation is that we only investigated relatively well-studied polymorphic variations in BDNF gene. Another limitation is that family-based studies and genome-wide association studies were excluded due to the heterogeneity of research methods. In a previous meta-analysis, the pooled OR derived from nine case–control studies is nominally significant (random-effects pooled OR = 1.07, P = 0.04), while the pooled OR derived from five family-based studies increases notably to an OR of 1.54 (P = 0.000019). Finally, since we primarily designed the current analysis to demonstrate the potential association(s) between Val66Met polymorphism and bipolar diagnostic boundaries, the association of this polymorphism with other characteristics of BPD such as sex, onset age, comorbidity, impairment in brain morphology and function, or treatment response, were not explored.
Conclusions
Taken together, the Val66Met polymorphism in BDNF gene may be involved in the pathogenesis of BPD by influencing the susceptibility of specific subtypes such as BP II, but there is no compelling evidence of BDNF gene playing an important role in susceptibility to BPD across different ethnicities. The associations observed in current meta-analysis should be interpreted with caution. Further large-scale studies with same definitions of phenotypes and controls in homogenous ethnic groups are warranted to elucidate the relevance of BDNF gene variations as a risk factor for BPD (or diagnostic subtypes) susceptibility.
References
Merikangas KR, Akiskal HS, Angst J, Greenberg PE, Hirschfeld RM, Petukhova M, Kessler RC: Lifetime and 12-month prevalence of bipolar spectrum disorder in the National Comorbidity Survey replication. Arch Gen Psychiatry. 2007, 64: 543-552. 10.1001/archpsyc.64.5.543.
Merikangas KR, Jin R, He JP, Kessler RC, Lee S, Sampson NA, Viana MC, Andrade LH, Hu C, Karam EG, Ladea M, Medina-Mora ME, Ono Y, Posada-Villa J, Sagar R, Wells JE, Zarkov Z: Prevalence and correlates of bipolar spectrum disorder in the world mental health survey initiative. Arch Gen Psychiatry. 2011, 68: 241-251. 10.1001/archgenpsychiatry.2011.12.
Phillips MR, Zhang J, Shi Q, Song Z, Ding Z, Pang S, Li X, Zhang Y, Wang Z: Prevalence, treatment, and associated disability of mental disorders in four provinces in China during 2001–05: an epidemiological survey. Lancet. 2009, 373: 2041-2053. 10.1016/S0140-6736(09)60660-7.
Geddes JR, Miklowitz DJ: Treatment of bipolar disorder. Lancet. 2013, 381: 1672-1682. 10.1016/S0140-6736(13)60857-0.
Yatham LN, Kennedy SH, Parikh SV, Schaffer A, Beaulieu S, Alda M, O’Donovan C, Macqueen G, McIntyre RS, Sharma V, Ravindran A, Young LT, Milev R, Bond DJ, Frey BN, Goldstein BI, Lafer B, Birmaher B, Ha K, Nolen WA, Berk M: Canadian Network for Mood and Anxiety Treatments (CANMAT) and International Society for Bipolar Disorders (ISBD) collaborative update of CANMAT guidelines for the management of patients with bipolar disorder: update 2013. Bipolar Disord. 2013, 15: 1-44. 10.1111/bdi.12025.
Muzina DJ, Kemp DE, McIntyre RS: Differentiating bipolar disorders from major depressive disorders: treatment implications. Ann Clin Psychiatry. 2007, 19: 305-312. 10.1080/10401230701653591.
Gao K, Kemp DE, Conroy C, Ganocy SJ, Findling RL, Calabrese JR: Comorbid anxiety and substance use disorders associated with a lower use of mood stabilizers in patients with rapid cycling bipolar disorder: a descriptive analysis of the cross-sectional data of 566 patients. Int J Clin Prac. 2010, 64: 336-344. 10.1111/j.1742-1241.2009.02284.x.
Hu C, Xiang YT, Ungvari GS, Dickerson FB, Kilbourne AM, Si TM, Fang YR, Lu Z, Yang HC, Chiu HF, Lai KY, Hu J, Chen ZY, Huang Y, Sun J, Wang XP, Li HC, Zhang JB, Wang G: Undiagnosed bipolar disorder in patients treated for major depression in China. J Affect Disord. 2012, 140: 181-186. 10.1016/j.jad.2012.02.014.
Xiang YT, Zhang L, Wang G, Hu C, Ungvari GS, Dickerson FB, Kilbourne AM, Si TM, Fang YR, Lu Z, Yang HC, Lai KY, Lee EH, Hu J, Chen ZY, Huang Y, Sun J, Wang XP, Li HC, Zhang JB, Chiu HF: Sociodemographic and clinical features of bipolar disorder patients misdiagnosed with major depressive disorder in China. Bipolar Disord. 2013, 15: 199-205. 10.1111/bdi.12052.
Wu R, Fan J, Zhao J, Calabrese JR, Gao K: The relationship between neurotrophins and bipolar disorder. Expert Rev Neurother. 2014, 14: 51-65. 10.1586/14737175.2014.863709.
Craddock N, Jones I: Genetics of bipolar disorder. J Med Genet. 1999, 36: 585-594.
McGuffin P, Rijsdijk F, Andrew M, Sham P, Katz R, Cardno A: The heritability of bipolar affective disorder and the genetic relationship to unipolar depression. Arch Gen Psychiatry. 2003, 60: 497-502. 10.1001/archpsyc.60.5.497.
Smoller JW, Finn CT: Family, twin, and adoption studies of bipolar disorder. Am J Med Genet C Semin Med Genet. 2003, 123C: 48-58. 10.1002/ajmg.c.20013.
Hayden EP, Nurnberger JI: Molecular genetics of bipolar disorder. Genes Brain Behav. 2006, 5: 85-95. 10.1111/j.1601-183X.2005.00138.x.
Fan J, Sklar P: Genetics of bipolar disorder: focus on BDNF Val66Met polymorphism. Novartis Found Symp. 2008, 289: 60-72. 10.1002/9780470751251.ch5.
Duman RS, Malberg J, Nakagawa S, D’Sa C: Neuronal plasticity and survival in mood disorders. Biol Psychiatry. 2000, 48: 732-739. 10.1016/S0006-3223(00)00935-5.
Manji HK, Moore GJ, Rajkowska G, Chen G: Neuroplasticity and cellular resilience in mood disorders. Mol Psychiatry. 2000, 5: 578-593. 10.1038/sj.mp.4000811.
Craddock N, Lendon C: Chromosome workshop: chromosomes 11, 14, and 15. Am J Med Genet. 1999, 88: 244-254. 10.1002/(SICI)1096-8628(19990618)88:3<244::AID-AJMG7>3.0.CO;2-1.
Cargill M, Altshuler D, Ireland J, Sklar P, Ardlie K, Patil N, Shaw N, Lane CR, Lim EP, Kalyanaraman N, Nemesh J, Ziaugra L, Friedland L, Rolfe A, Warrington J, Lipshutz R, Daley GQ, Lander ES: Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat Genet. 1999, 22: 231-238. 10.1038/10290.
Neves-Pereira M, Mundo E, Muglia P, King N, Macciardi F, Kennedy JL: The brain-derived neurotrophic factor gene confers susceptibility to bipolar disorder: evidence from a family-based association study. Am J Hum Genet. 2002, 71: 651-655. 10.1086/342288.
Sklar P, Gabriel SB, McInnis MG, Bennett P, Lim Y, Tsan G, Schaffner S, Kirov G, Jones I, Owen M, Craddock N, DePaulo JR, Lander ES: Family-based association study of 76 candidate genes in bipolar disorder: BDNF is a potential risk locus. brain-derived neutrophic factor. Mol Psychiatry. 2002, 7: 579-593. 10.1038/sj.mp.4001058.
Kanazawa T, Glatt SJ, Kia-Keating B, Yoneda H, Tsuang MT: Meta-analysis reveals no association of the Val66Met polymorphism of brain-derived neurotrophic factor with either schizophrenia or bipolar disorder. Psychiatr Genet. 2007, 17: 165-170. 10.1097/YPG.0b013e32801da2e2.
Dersimonian R: Combining evidence from clinical-trials. Anesth Analg. 1990, 70: 475-476. 10.1213/00000539-199005000-00001.
Woolf B: On estimating the relation between blood group and disease. Ann Hum Genet. 1955, 19: 251-253. 10.1111/j.1469-1809.1955.tb01348.x.
The Cochrane Collaboration: Review Manager (RevMan) [Computer program] Version 5.2. 2012, The Nordic Cochrane Centre, Copenhagen
Huang CC, Chang YH, Lee SY, Chen SL, Chen SH, Chu CH, Huang SY, Tzeng NS, Lee IH, Yeh TL, Yang YK, Lu RB: The interaction between BDNF and DRD2 in bipolar II disorder but not in bipolar I disorder. Am J Med Genet B Neuropsychiatr Genet. 2012, 159B: 501-507. 10.1002/ajmg.b.32055.
Lee SY, Chen SL, Chen SH, Chu CH, Chang YH, Lin SH, Huang SY, Tzeng NS, Kuo PH, Lee IH, Yeh TL, Yang YK, Lu RB: Interaction of the DRD3 and BDNF gene variants in subtyped bipolar disorder. Prog Neuropsychopharmacol Biol Psychiatry. 2012, 39: 382-387. 10.1016/j.pnpbp.2012.07.015.
Lee SY, Chen SL, Wang YS, Chang YH, Huang SY, Tzeng NS, Lee IH, Yeh TL, Yang YK, Lu RB: COMT and BDNF interacted in bipolar II disorder not comorbid with anxiety disorder. Behav Brain Res. 2013, 237: 243-248. 10.1016/j.bbr.2012.09.039.
Chen SL, Lee SY, Chang YH, Chen SH, Chu CH, Wang TY, Chen PS, Lee IH, Yang YK, Hong JS, Lu RB: The BDNF Val66Met polymorphism and plasma brain-derived neurotrophic factor levels in Han Chinese patients with bipolar disorder and schizophrenia. Prog Neuropsychopharmacol Biol Psychiatry. 2014, 51C: 99-104. 10.1016/j.pnpbp.2014.01.012.
Chang YH, Lee SY, Chen SL, Tzeng NS, Wang TY, Lee IH, Chen PS, Huang SY, Yang YK, Ko HC, Lu RB: Genetic variants of the BDNF and DRD3 genes in bipolar disorder comorbid with anxiety disorder. J Affect Disord. 2013, 151: 967-972. 10.1016/j.jad.2013.08.017.
Chepenik LG, Fredericks C, Papademetris X, Spencer L, Lacadie C, Wang F, Pittman B, Duncan JS, Staib LH, Duman RS, Gelernter J, Blumberg HP: Effects of the brain-derived neurotrophic growth factor val66met variation on hippocampus morphology in bipolar disorder. Neuropsychopharmacology. 2009, 34: 944-951. 10.1038/npp.2008.107.
Matsuo K, Walss-Bass C, Nery FG, Nicoletti MA, Hatch JP, Frey BN, Monkul ES, Zunta-Soares GB, Bowden CL, Escamilla MA, Soares JC: Neuronal correlates of brain-derived neurotrophic factor Val66Met polymorphism and morphometric abnormalities in bipolar disorder. Neuropsychopharmacology. 2009, 34: 1904-1913. 10.1038/npp.2009.23.
Carrard A, Salzmann A, Perrou N, Gafner J, Malafoss A, Karege F: Genetic association of the Phosphoinositide-3 kinase in schizophrenia and bipolar disorder and interaction with a BDNF gene polymorphism. Brain Behav. 2011, 1: 119-124. 10.1002/brb3.23.
Gruber O, Hasan A, Scherk H, Wobrock T, Schneider-Axmann T, Ekawardhani S, Schmitt A, Backens M, Reith W, Meyer J, Falkai P: Association of the brain-derived neurotrophic factor val66met polymorphism with magnetic resonance spectroscopic markers in the human hippocampus: in vivo evidence for effects on the glutamate system. Eur Arch Psychiatry Clin Neurosci. 2012, 262: 23-31. 10.1007/s00406-011-0214-6.
Teh CA, Lee TS, Kuchibhatla M, Ashley-Koch A, Macfall J, Krishnan R, Beyer J: Bipolar disorder, brain-derived neurotrophic factor (BDNF) Val66Met polymorphism and brain morphology. PLoS One. 2012, 7: e38469-10.1371/journal.pone.0038469.
Kenna HA, Reynolds-May M, Stepanenko A, Ketter TA, Hallmayer J, Rasgon NL: Blood levels of brain derived neurotrophic factor in women with bipolar disorder and healthy control women. J Affect Disord. 2014, 156: 214-218. 10.1016/j.jad.2013.01.054.
Wellcome Trust Case Control Consortium: Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007, 447: 661-678. 10.1038/nature05911.
Hong CJ, Huo SJ, Yen FC, Tung CL, Pan GM, Tsai SJ: Association study of a brain-derived neurotrophic-factor genetic polymorphism and mood disorders, age of onset and suicidal behavior. Neuropsychobiology. 2003, 48: 186-189. 10.1159/000074636.
Nakata K, Ujike H, Sakai A, Uchida N, Nomura A, Imamura T, Katsu T, Tanaka Y, Hamamura T, Kuroda S: Association study of the brain-derived neurotrophic factor (BDNF) gene with bipolar disorder. Neurosci Lett. 2003, 337: 17-20. 10.1016/S0304-3940(02)01292-2.
Kunugi H, Iijima Y, Tatsumi M, Yoshida M, Hashimoto R, Kato T, Sakamoto K, Fukunaga T, Inada T, Suzuki T, Iwata N, Ozaki N, Yamada K, Yoshikawa T: No association between the Val66Met polymorphism of the brain-derived neurotrophic factorgene and bipolar disorder in a Japanese population: a multicenter study. Biol Psychiatry. 2004, 56: 376-378. 10.1016/j.biopsych.2004.06.017.
Oswald P, Del-Favero J, Massat I, Souery D, Claes S, Van Broeckhoven C, Mendlewicz J: Non-replication of the brain-derived neurotrophic factor (BDNF) association in bipolar affective disorder: a Belgian patient-control study. Am J Med Genet B Neuropsychiatr Genet. 2004, 129B: 34-35. 10.1002/ajmg.b.30056.
Skibinska M, Hauser J, Czerski PM, Leszczynska-Rodziewicz A, Kosmowska M, Kapelski P, Slopien A, Zakrzewska M, Rybakowski JK: Association analysis of brain-derived neurotrophic factor (BDNF) gene Val66Met polymorphism in schizophrenia and bipolar affective disorder. World J Biol Psychiatry. 2004, 5: 215-220. 10.1080/15622970410029936.
Lohoff FW, Sander T, Ferraro TN, Dahl JP, Gallinat J, Berrettini WH: Confirmation of association between the Val66Met polymorphism in the brain-derived neurotrophic factor (BDNF) gene and bipolar I disorder. Am J Med Genet B Neuropsychiatr Genet. 2005, 139B: 51-53. 10.1002/ajmg.b.30215.
Neves-Pereira M, Cheung JK, Pasdar A, Zhang F, Breen G, Yates P, Sinclair M, Crombie C, Walker N, St Clair DM: BDNF gene is a risk factor for schizophrenia in a Scottish population. Mol Psychiatry. 2005, 10: 208-212. 10.1038/sj.mp.4001575.
Schumacher J, Jamra RA, Becker T, Ohlraun S, Klopp N, Binder EB, Schulze TG, Deschner M, Schmäl C, Höfels S, Zobel A, Illig T, Propping P, Holsboer F, Rietschel M, Nöthen MM, Cichon S: Evidence for a relationship between genetic variants at the brain-derived neurotrophic factor (BDNF) locus and major depression. Biol Psychiatry. 2005, 58: 307-314. 10.1016/j.biopsych.2005.04.006.
Green EK, Raybould R, Macgregor S, Hyde S, Young AH, O’Donovan MC, Owen MJ, Kirov G, Jones L, Jones I, Craddock N: Genetic variation of brain-derived neurotrophic factor (BDNF) in bipolar disorder: case–control study of over 3000 individuals from the UK. Br J Psychiatry. 2006, 188: 21-25. 10.1192/bjp.bp.105.009969.
Liu M, Ling SH, Li WB, Wang CY, Chen DF, Wang G: An association study between GRIN1, BDNF genes and bipolar disorder. Yi Chuan. 2007, 29: 41-46. 10.1360/yc-007-0041.
Tramontina J, Frey BN, Andreazza AC, Zandona M, Santin A, Kapczinski F: Val66met polymorphism and serum brain-derived neurotrophic factor levels in bipolar disorder. Mol Psychiatry. 2007, 12: 230-231. 10.1038/sj.mp.4001941.
Kim B, Kim CY, Hong JP, Kim SY, Lee C, Joo YH: Brain-derived neurotrophic factor Val/Met polymorphism and bipolar disorder. association of the Met allele with suicidal behavior of bipolar patients. Neuropsychobiology. 2008, 58: 97-103. 10.1159/000162356.
Tang J, Xiao L, Shu C, Wang G, Liu Z, Wang X, Wang H, Bai X: Association of the brain-derived neurotrophic factor gene and bipolar disorder with early age of onset in mainland China. Neurosci Lett. 2008, 433: 98-102. 10.1016/j.neulet.2008.01.001.
Vincze I, Perroud N, Buresi C, Baud P, Bellivier F, Etain B, Fournier C, Karege F, Matthey ML, Preisig M, Leboyer M, Malafosse A: Association between brain-derived neurotrophic factor gene and a severe form of bipolar disorder, but no interaction with the serotonin transporter gene. Bipolar Disord. 2008, 10: 580-587. 10.1111/j.1399-5618.2008.00603.x.
Ye CY, Xu YQ, Hu H, Yu SY, Wang DX, Shi SX, Wang LW: An association study of brain-derived neurotrophic factor gene polymorphism in bipolar disorders. Zhonghua Yi Xue Za Zhi. 2009, 89: 1897-1901.
Hosang GM, Uher R, Keers R, Cohen-Woods S, Craig I, Korszun A, Perry J, Tozzi F, Muglia P, McGuffin P, Farmer AE: Stressful life events and the brain-derived neurotrophic factor gene in bipolar disorder. J Affect Disord. 2010, 125: 345-349. 10.1016/j.jad.2010.01.071.
Xu J, Liu Y, Wang P, Li S, Wang Y, Li J, Zhou D, Chen Z, Zhao T, Wang T, Xu H, Yang Y, Feng G, He L, Yu L: Positive association between the brain-derived neurotrophic factor (BDNF) gene and bipolar disorder in the Han Chinese population. Am J Med Genet B Neuropsychiatr Genet. 2010, 153B: 275-279. 10.1002/ajmg.b.31096.
Min HJ, Cho HS, Kim SJ, Seok JH, Lee E, Jon DI: Association of the brain-derived neurotrophic factor gene and clinical features of bipolar disorder in Korea. Clin Psychopharmacol Neurosci. 2012, 10: 163-167. 10.9758/cpn.2012.10.3.163.
Wang Z, Li Z, Chen J, Huang J, Yuan C, Hong W, Yu S, Fang Y: Association of BDNF gene polymorphism with bipolar disorders in Han Chinese population. Genes Brain Behav. 2012, 11: 524-528. 10.1111/j.1601-183X.2012.00797.x.
Pae CU, Chiesa A, Porcelli S, Han C, Patkar AA, Lee SJ, Park MH, Serretti A, De Ronchi D: Influence of BDNF variants on diagnosis and response to treatment in patients with major depression, bipolar disorder and schizophrenia. Neuropsychobiology. 2012, 65: 1-11. 10.1159/000327605.
Petryshen TL, Sabeti PC, Aldinger KA, Fry B, Fan JB, Schaffner SF, Waggoner SG, Tahl AR, Sklar P: Population genetic study of the brain-derived neurotrophic factor (BDNF) gene. Mol Psychiatry. 2010, 15: 810-815. 10.1038/mp.2009.24.
Müller DJ, de Luca V, Sicard T, King N, Strauss J, Kennedy JL: Brain-derived neurotrophic factor (BDNF) gene and rapid-cycling bipolar disorder: family-based association study. Br J Psychiatry. 2006, 189: 317-323. 10.1192/bjp.bp.105.010587.
Liu L, Foroud T, Xuei X, Berrettini W, Byerley W, Coryell W, El-Mallakh R, Gershon ES, Kelsoe JR, Lawson WB, MacKinnon DF, McInnis M, McMahon FJ, Murphy DL, Rice J, Scheftner W, Zandi PP, Lohoff FW, Niculescu AB, Meyer ET, Edenberg HJ, Nurnberger JI: Evidence of association between brain-derived neurotrophic factor gene and bipolar disorder. Psychiatr Genet. 2008, 18: 267-274. 10.1097/YPG.0b013e3283060f59.
Acknowledgements
Funding for this study was provided by the National Natural Science Foundation of China (Grant# 81301159, 91232719), Shanghai Key Medicine Specialties Program (Grant# ZK2012A12), Training Plan for Excellent Academic Leaders of Shanghai Health System (Grant# XBR2013087), the “12th Five-year Plan” of National Key Technologies R&D Program (Grant# 2012BAI01B04), and the National Key Clinical Disciplines at Shanghai Mental Health Center (Grant# OMA-MH 2011–873). The authors thank Dr Lihua Zhi, Academic Communication Officer in Eli Lilly Asia, Inc., who provided kind help in searching the literatures.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
ZW, KG and YF contributed to conception, design, analysis and interpretation of data and drafting of the manuscript. ZW and ZL reviewed all references and extracted the data from each eligible study. All authors have given final approval of the version to be published.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Wang, Z., Li, Z., Gao, K. et al. Association between brain-derived neurotrophic factor genetic polymorphism Val66Met and susceptibility to bipolar disorder: a meta-analysis. BMC Psychiatry 14, 366 (2014). https://doi.org/10.1186/s12888-014-0366-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12888-014-0366-9