Abstract
Objectives
We performed an updated meta-analysis to clarify the relationship between the CEBPE rs2239633 polymorphism and the childhood acute lymphoblastic leukemia (CALL) susceptibility.
Methods
All the case-control studies were updated on October 5, 2020, through Web of Science, PubMed, Cochrane Library, Embase, and China National Knowledge Infrastructure (CNKI) electronic database. The heterogeneity in the study was tested by the Q test and I2, and then the random ratio or fixed effect was utilized to merge the odds ratios (OR) and 95% confidence interval (CI). We also performed sensitivity analysis to estimate the impact of individual studies on aggregate estimates. Publication bias was investigated by using funnel plot and Egger’s regression test. All statistical analyses were performed using Stata 12.0.
Results
A total of 20 case-control studies were selected, including 7014 patients and 16,428 controls. There was no association of CEBPE rs2239633 polymorphism with CALL (CC vs CT + TT: OR = 1.08, 95% CI = 0.94–1.26; CC + CT vs TT: OR = 1.10, 95% CI = 0.94–1.30; C vs T: OR = 1.02, 95% CI = 0.92–1.13). In the subgroup analysis by ethnicity, there is no significant association of this polymorphism and CALL risks among Asian and Caucasian populations in the three genetic models (CC vs CT + TT, CC + CT vs TT, and C vs T).
Conclusion
This meta-analysis found no significant association between the CEBPE rs2239633 polymorphism and susceptibility to CALL.
Similar content being viewed by others
Introduction
Acute lymphoblastic leukemia (ALL) is a malignant disease of the blood system. It occurs mostly in children under 15 years of age. The peak age of onset was 2–5 years old [1, 2], accounting for about 1/3 of childhood malignant tumors [3]. Although the etiology and pathogenesis were not yet clear, previous studies had shown that ALL was the result of multiple factors such as genetic variation and exposure to carcinogens in the environment [4, 5]. In recent years, genome-wide association studies (GWAS) had shown that gene mononucleotide polymorphism (SNP) variation was an important risk factor for CALL [6,7,8,9].
The CEBPE gene was located on the human chromosome 14q11.2, which was a member of the CCAAT-enhancer-binding protein family, and its encoded protein belongs to the basic leucine transcription factor. The CEBPE gene-encoded protein was essential for terminal differentiation and functional maturation of myeloid committed progenitor cells, especially for the maturation of neutrophils and giant wah cells [10]. Mutations in CEBPE would cause loss of neutrophil granules [11]. Akasaka had reported that CEBPE mutations can cause translocation of immunoglobulin heavy chain chromosomes, which often occurred in children with B-precursor E cell leukemia [12]. This indicated that the CEBPE gene played an important role in the occurrence and development of ALL.
Two meta-analysis studies in 2014 [13] and 2015 [14] found the association of CEBPE rs2239633 polymorphism with the risk of CALL, but the conclusions obtained from the two studies were reversed. In addition, since 2015, many studies had reported CEBPE rs2239633 polymorphisms and the risk of CALL [11, 15,16,17,18,19]. Therefore, the purpose of this meta-analysis was to investigate the relationship between CEBPE rs2239633 polymorphism and the risk of CALL.
Materials and methods
Search strategies
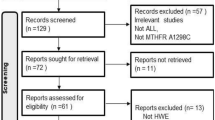
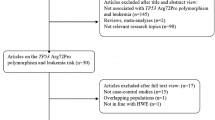
We conducted a systematic online search of the literature in the Web of Science, PubMed, Cochrane Library, Embase, and China National Knowledge Infrastructure (CNKI) electronic database, covering relevant studies published until October 5, 2020. The keywords for the search were as follows: (“rs2239633” OR “CEBPE”) AND (“polymorphism” OR “variant” OR “mutation”) AND (“acute lymphoblastic leukemia” or “ALL”). The literature on relevant data was searched in English and Chinese, respectively. In addition, the retrieved articles and references were performed with manual searches. Referring to the Preferred Reporting Project (PRISMA) Guide for Systematic Evaluation and Meta-Analysis [20], an information flow diagram related to the final eligibility data was constructed by screening all retrieved literature.
Inclusion and exclusion criteria
Screening for the studies of the relationship between CEBPE rs2239633 polymorphism and the risk of ALL is according to the following inclusion criteria: (1) the design of the study was case-control, (2) the full text can be found, (3) the genotype information of the CEBPE rs2239633 polymorphism was available, and (4) the relationship of the CEBPE rs2239633 polymorphism and the risk of ALL was evaluated. The major exclusion criteria were (1) not a case-control study; (2) repeating early publications (studies used in different publications for the same sample data, including only the most complete samples after careful review); (3) unpublished articles, conference papers, meta-analysis, and systematic reviews; and (4) family-based pedigree research. This meta-analysis strictly followed the requirements of the preferred reporting project for the systematic review and meta-analysis guidelines [20].
Data extraction
The analysis data of the selected studies were independently extracted by two researchers using standard data collection forms. Study-related information extracted from each literature was as follows: first author, year of publication, country of origin, mean age and gender in cases and controls, numbers of cases and controls, Hardy-Weinberg equilibrium, genotyping method, source of controls, and available genotype frequency information for CEBPE rs2239633. If the same sample data appeared in multiple publications, only the publication with the largest sample size was included in the study. The differences between the two investigators were resolved through discussion. If the discussion could not resolve the objection between the two, the objection would be judged by the third investigator. All data were obtained from the full text of the published research, and the author was not contacted for further information.
Study quality assessment
Two evaluators evaluated the quality of the included studies according to the Newcastle-Ottawa Scale (NOS) [21], which was applicable to the quality assessment of observational studies. The difference between the two evaluators was reported and resolved by the third evaluator. The scores of research quality mainly included the following three aspects: (1) selection of the case groups and control groups (4 stars), (2) quality of confounding factor correction in case and control population (2 stars), and (3) determination of the exposure of interest in the studies (3 stars). For each item numbered in the selection and exposure categories, one study can be rated as up to one star, and comparability can be assigned up to two stars. Higher scores indicate an increase in the quality of the research method. Studies with scores equal to or higher than 6 are considered high-quality studies.
Data analysis
The heterogeneity in the study was tested by the Q test and I2 [22, 23], and then the random ratio or fixed effect was utilized to merge the odds ratios (OR) and 95% confidence interval (CI) [24]. The significance of the pooled OR was analyzed by Z test (P < 0.05 judged statistically significant). To estimate the impact of individual studies on aggregate estimates, we also performed sensitivity analysis [25]. Using funnel plot and Eegger’s regression test investigated the publication bias [26, 27]. All data statistical analyses were performed using Stata 12.0 (Stata Corp, College Station, TX, USA).
Results
Literature search and study characteristics
The flow chart of the literature search was shown in Fig. 1. One hundred sixty-five potentially relevant articles were selected in the preliminary online search. After verifying and deleting 80 duplicate articles, 85 articles entered the final review. Through the review of the title and abstract, 26 articles were included for full-text review. Finally, 16 articles were included in the final study. These studies were published between 2009 and 2017, and 20 studies included 7014 ALL patients and 16,428 controls. The distribution of genotypes in controls in all studies followed HWE. In addition, the NOS scores for all studies ranged from 6 to 8 points, so that the selected articles were considered to be good in methodological quality. The relevant feature information of the included articles was in Tables 1 and 2.
Meta-analysis results
The heterogeneity of the three genetic models was determined by Q test and I2 statistics. As shown in Fig. 2, these were serious heterogeneity in the three models (CC vs CT + TT: P < 0.001, I2 = 63.6%; CC + CT vs TT: P = 0.002, I2 = 70.2%; C vs T: P < 0.001, I2 = 79.2%); thus, we used the random-effect model to analyze the three models. Our results did not find significant associations between CEBPE rs2239633 polymorphism and the risk of ALL under the three models, CC vs CT + TT (OR = 1.08, 95% CI = 0.94–1.26, P = 0.280), CC + CT vs TT (OR = 1.10, 95% CI = 0.94–1.30, P = 0.228), and C vs T (OR = 1.02, 95% CI = 0.92–1.13, P = 0.752). In the subgroup analysis by ethnicity, no significant association was found in three models in both Caucasian and Asian populations (Table 3). Sensitivity analysis was used to assess the impact of each individual study on the pooled OR by sequentially removing each eligible study. Our results suggest that none of the studies affected the overall outcome of the pooled OR (Fig. 3). Begg’s funnel plot was used to assess publication bias, and the results showed that publication bias was not reflected in the three genetic models (CC vs CT + TT: P = 0.742; CC + CT vs TT: P = 0.285; C vs T: P = 0.560) (Fig. 4).
Discussion
As a transcription factor specifically expressed in myeloid cells, CCAAT/enhancer-binding protein-ε (CEBPE) played an important role in the proliferation, growth, differentiation, and apoptosis of myeloid cells and participates in the transcriptional regulation of a series of myeloid-specific genes. Loss of activity was an important factor leading to the onset of bone marrow disease [28]. In recent years, a growing number of published studies had investigated the relationship between CEBPE rs2239633 polymorphism and ALL risk [29,30,31,32,33,34,35,36,37]. It also included some meta-analysis, but the results obtained from the meta-analysis were contradictory and conflicting. To further assess the relationship between CEBPE rs2239633 polymorphism and ALL risk, we performed an updated meta-analysis to investigate the relationship between CEBPE rs2239633 polymorphism and ALL risk.
Although the GWAS study by Papaemmanuil et al. [6] proved that the 5′ SNP rs2239633 located in CEBPE has a strong correlation with children’s ALL in the European population, however, this meta-analysis showed that no significant association was found in the three selected genetic models. In the subgroup analysis of ethnicity, no significant correlation was found under the three genetic models. On the one hand, this difference may be caused by the linkage imbalance between populations. There are also some differences between the population samples. On the other hand, the exact pathogenesis of CEBPE in the etiology of leukemia was still unclear. The CEBPE mutation may have different effects on the immune system of different children.
Previously, a meta-analysis was performed for 11 case-control studies with 5639 cases and 10,036 controls by Wang et al. [13], the results showed no association of the CEBPE rs2239633 polymorphism and childhood ALL risk, and subgroup analysis stratified by ethnicity found a significant association of this polymorphism with childhood ALL in the Caucasian subgroup and Hispanic subgroup, but not in the Asian subgroup. Sun et al. [14] also conducted a meta-analysis, including 22 published studies involving 6152 patients and 11,739 healthy controls, and the results also showed that CEBPE rs2239633 variant was associated with decreased risk of childhood B cell ALL in Europeans, but not among T cell ALL, Asian, and mixed populations. The results of the two meta-analyses are diametrically opposed, and this difference may be due to the difference in the number of samples included and the sample size. This study combines the latest research literature with the first two meta-analyses to more fully describe the relationship between CEBPE rs2239633 and CALL. In terms of statistical power, it is significantly better than the previous meta-analysis of Sun et al. [14] and Wang et al. [13].
However, there are certain limitations in our research. First, databases that include only published research in both Chinese and English are selected for analysis, and other languages or unpublished potential researches may be missed. Second, due to the lack of raw data, we were unable to assess potential interactions of gene-genes and genes-environments. Third, the meta-analysis includes data from Europeans and Asians, so the results of this item apply only to these two ethnic groups. Fourth, among the three models, heterogeneity may greatly influence the conclusion of the meta-analysis. Lastly, maybe the results obtained from this study have limited clinic significance, but we think that the current study was needed and meaningful for understanding the relationship between the CEBPE rs2239633 polymorphism and the CALL susceptibility.
Conclusion
In summary, our study showed that the CEBPE rs2239633 gene polymorphism did not increase or decrease the risk of susceptibility to CALL. Although the specific causes of childhood leukemia were still unclear, a large number of existing researches tended to suggest that the occurrence of childhood ALL was the result of a combination of factors, especially the genetic and environmental factors. Therefore, in the future, when studying the relationship between CEBPE rs2239633 polymorphism and childhood ALL, the influence of environmental factors on the relationship between the two should be removed.
Availability of data and materials
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Eden T. Aetiology of childhood leukaemia. Cancer Treat Rev. 2010;36(4):286–97.
Terracini B. Epidemiology of childhood cancer. Environ Health 2011; 10 Suppl 1(Suppl 1):S8.
Karathanasis NV, Choumerianou DM, Kalmanti M. Gene polymorphisms in childhood ALL. Pediatr Blood Cancer. 2008;52(3):318–23.
Bhojwani D, Yang JJ, Pui CH. Biology of childhood acute lymphoblastic leukemia. Pediatr Clin N Am. 2015;62(1):47–60.
Schuz J, Erdmann F. Environmental exposure and risk of childhood leukemia: an overview. Arch Med Res. 2016;47(8):607–14.
Papaemmanuil E, Hosking FJ, Vijayakrishnan J, Price A, Olver B, Sheridan E, et al. Loci on 7p12.2, 10q21.2 and 14q11.2 are associated with risk of childhood acute lymphoblastic leukemia. Nat Genet. 2009;41(9):1006–10.
TrevnO LR, Yang W, French D, Hunger SP, Carroll WL, Devidas M, et al. Germline genomic variants associated with childhood acute lymphoblastic leukemia. Nat Genet. 2009;41(9):1001–5.
Orsi L, Rudant J, Bonaventure A, Goujon-Bellec S, Corda E, Evans TJ, et al. Genetic polymorphisms and childhood acute lymphoblastic leukemia: GWAS of the ESCALE study (SFCE). Leukemia. 2012;26(12):2561–4.
Walsh KM, Chokkalingam AP, Hsu LI, Metayer C, de Smith AJ, Jacobs DI, et al. Associations between genome-wide native American ancestry, known risk alleles and B-cell ALL risk in Hispanic children. Leukemia. 2013;27(12):2416–9.
Akagi T, Thoennissen NH, George A, Crooks G, Song JH, Okamoto R, et al. In vivo deficiency of both C/EBPbeta and C/EBPepsilon results in highly defective myeloid differentiation and lack of cytokine response. PLoS One. 2010;5(11):e15419.
Gharbi H, Ben Hassine I, Soltani I, Safra I, Ouerhani S, Bel Haj Othmen H, et al. Association of genetic variation in IKZF1, ARID5B, CDKN2A, and CEBPE with the risk of acute lymphoblastic leukemia in Tunisian children and their contribution to racial differences in leukemia incidence. Pediatr Hematol Oncol. 2016;33(3):157–67.
Akasaka T, Balasas T, Russell LJ, Sugimoto KJ, Majid A, Walewska R, et al. Five members of the CEBP transcription factor family are targeted by recurrent IGH translocations in B-cell precursor acute lymphoblastic leukemia (BCP-ALL). Blood. 2007;109(8):3451–61.
Wang C, Chen J, Sun H, Sun L, Liu Y. CEBPE polymorphism confers an increased risk of childhood acute lymphoblastic leukemia: a meta-analysis of 11 case-control studies with 5,639 cases and 10,036 controls. Ann Hematol. 2015;94(2):181–5.
Sun J, Zheng J, Tang L, Healy J, Sinnett D, Dai YE. Association between CEBPE variant and childhood acute leukemia risk: evidence from a meta-analysis of 22 studies. PLoS One. 2015;10(5):e0125657.
Al-Absi B, Razif MFM, Noor SM, Saif-Ali R, Aqlan M, Salem SD, et al. Contributions of IKZF1, DDC, CDKN2A, CEBPE, and LMO1 gene polymorphisms to acute lymphoblastic leukemia in a Yemeni population. Genetic Test Mol Biomark. 2017;21(10):592–9.
Bekker-Mendez VC, Nunez-Enriquez JC, Torres Escalante JL, Alvarez-Olmos E, Gonzalez-Montalvoc PM, Jimenez-Hernandez E, et al. ARID5B, CEBPE and PIP4K2A germline genetic polymorphisms and risk of childhood acute lymphoblastic leukemia in Mexican patients: a MIGICCL study. Arch Med Res. 2016;47(8):623–8.
Bhandari P, Ahmad F, Mandava S, Das BR. Association of genetic variants in ARID5B, IKZF1 and CEBPE with risk of childhood de novo B-lineage acute lymphoblastic leukemia in India. Asian Pac J Cancer Prev. 2016;17(8):3989–95.
Urayama KY, Takagi M, Kawaguchi T, Matsuo K, Tanaka Y, Ayukawa Y, et al. Regional evaluation of childhood acute lymphoblastic leukemia genetic susceptibility loci among Japanese. Sci Rep. 2018;8(1):789.
Kreile M, Piekuse L, Rots D, Dobele Z, Kovalova Z, Lace B. Analysis of possible genetic risk factors contributing to development of childhood acute lymphoblastic leukaemia in the Latvian population. Arch Med Sci. 2016;12(3):479–85.
Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med. 2009;151(4):264–9 w264.
Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25(9):603–5.
Higgins JPT, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 21(11):1539–58.
Zintzaras E, Lau J. Synthesis of genetic association studies for pertinent gene-disease associations requires appropriate methodological and statistical approaches. J Clin Epidemiol. 2008;61(7):634–45.
DerSimonian R, Laird N. Meta-analysis in clinical trials revisited. Contemp Clin Trials. 2015;45(Pt A):139–45.
Copas J. Meta-analysis, funnel plots and sensitivity analysis. Biostatistics. 2000;1(3):247–62.
Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ (Clinical research ed). 1997;315(7109):629–34.
Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50(4):1088–101.
Wada T, Akagi T. Role of the leucine zipper domain of CCAAT/enhancer binding protein-epsilon (C/EBPepsilon) in neutrophil-specific granule deficiency. Crit Rev Immunol. 2016;36(4):349–58.
Ellinghaus E, Stanulla M, Richter G, Ellinghaus D, te Kronnie G, Cario G, et al. Identification of germline susceptibility loci in ETV6-RUNX1-rearranged childhood acute lymphoblastic leukemia. Leukemia. 2012;26(5):902–9.
Prasad RB, Hosking FJ, Jayaram V, Elli P, Rolf K, Mel G, et al. Verification of the susceptibility loci on 7p12.2, 10q21.2, and 14q11.2 in precursor B-cell acute lymphoblastic leukemia of childhood. Blood. 2010;115(9):1765–7.
Vijayakrishnan J, Sherborne AL, Sawangpanich R, Hongeng S, Houlston RS, Pakakasama S. Variation at 7p12.2 and 10q21.2 influences childhood acute lymphoblastic leukemia risk in the Thai population and may contribute to racial differences in leukemia incidence. Leukemia Lymphoma. 2010;51(10):1870–4.
Pastorczak A, Górniak P, Sherborne A, Hosking F, Trelińska J, Lejman M, et al. Role of 657del5 NBN mutation and 7p12.2 (IKZF1), 9p21 (CDKN2A), 10q21.2 (ARID5B) and 14q11.2 (CEBPE) variation and risk of childhood ALL in the polish population. Leuk Res. 2011;35(11):1534–6.
Lautner-Csorba O, Gezsi A, Semsei AF, Antal P, Erdelyi DJ, Schermann G, et al. Candidate gene association study in pediatric acute lymphoblastic leukemia evaluated by Bayesian network based Bayesian multilevel analysis of relevance. BMC Med Genet. 2012;5:42.
Chokkalingam AP, Hsu LI, Metayer C, Hansen HM, Month SR, Barcellos LF, et al. Genetic variants in ARID5B and CEBPE are childhood ALL susceptibility loci in Hispanics. Cancer Causes Control. 2013;24(10):1789–95.
Ross JA, Linabery AM, Blommer CN, Langer EK, Spector LG, Hilden JM, et al. Genetic variants modify susceptibility to leukemia in infants: a Children’s oncology group report. Pediatr Blood Cancer. 2013;60(1):31–4.
Wang Y, Chen J, Li J, Deng J, Rui Y, Lu Q, et al. Association of three polymorphisms in ARID5B, IKZF1 and CEBPE with the risk of childhood acute lymphoblastic leukemia in a Chinese population. Gene. 2013;524(2):203–7.
Emerenciano M, Barbosa TC, Lopes BA, Blunck CB, Faro A, Andrade C, et al. ARID5B polymorphism confers an increased risk to acquire specific MLL rearrangements in early childhood leukemia. BMC Cancer. 2014;14:127.
Acknowledgements
We appreciate the cooperation of the partners and staff that cooperated in this study.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
Manuscript writing, editing, and reviewing were conducted by JL. GW and LX participated in the article search. WL and CZ performed data analysis and evaluated the quality of the selected studies. The authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Liu, J., Weiling, G., Xueqin, L. et al. The CEBPE rs2239633 genetic polymorphism on susceptibility to childhood acute lymphoblastic leukemia: an updated meta-analysis. Environ Health Prev Med 26, 2 (2021). https://doi.org/10.1186/s12199-020-00920-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12199-020-00920-2