Abstract
Background
Loricifera is an enigmatic metazoan phylum; its morphology appeared to place it with Priapulida and Kinorhyncha in the group Scalidophora which, along with Nematoida (Nematoda and Nematomorpha), comprised the group Cycloneuralia. Scarce molecular data have suggested an alternative phylogenetic hypothesis, that the phylum Loricifera is a sister taxon to Nematomorpha, although the actual phylogenetic position of the phylum remains unclear.
Methods
Ecdysozoan phylogeny was reconstructed through maximum-likelihood (ML) and Bayesian inference (BI) analyses of nuclear 18S and 28S rRNA gene sequences from 60 species representing all eight ecdysozoan phyla, and including a newly collected loriciferan species.
Results
Ecdysozoa comprised two clades with high support values in both the ML and BI trees. One consisted of Priapulida and Kinorhyncha, and the other of Loricifera, Nematoida, and Panarthropoda (Tardigrada, Onychophora, and Arthropoda). The relationships between Loricifera, Nematoida, and Panarthropoda were not well resolved.
Conclusions
Loricifera appears to be closely related to Nematoida and Panarthropoda, rather than grouping with Priapulida and Kinorhyncha, as had been suggested by previous studies. Thus, both Scalidophora and Cycloneuralia are a polyphyletic or paraphyletic groups. In addition, Loricifera and Nematomorpha did not emerge as sister groups.
Similar content being viewed by others
Introduction
Since its first description as a new phylum [1], Loricifera has been one of the most enigmatic metazoan phyla. Although only 35 loriciferan species have been described worldwide, the actual species diversity is higher, as many new species await description [2–6]. All known loriciferan species are microscopic (80–800 μm) and occur in marine sediments, such as mud, sand, and shell gravel. The most extreme habitat for Loricifera is the hypersaline anoxic deep basin in the Mediterranean Sea, where members of this phylum are metabolically active [6, 7]. Our knowledge of loriciferan life cycles is also only fragmentary, given the recent findings of new life cycles and larval types [3–5, 8].
There are two alternative hypotheses on the position of Loricifera within Ecdysozoa, both based on morphological data. One is the ‘Scalidophora hypothesis’ [9–11], in which Loricifera, Kinorhyncha, and Priapulida together comprise a clade, Scalidophora. Morphological similarities between Scalidophora and Nematomorpha [12–15] and between Scalidophora and Nematoida (Nematomorpha and Nematoda) [9, 11, 16–21] have indicated that these five phyla in turn comprise a clade, Cycloneuralia [20, 21].
The alternative is the ‘Loricifera + Nematomorpha hypothesis’ [22]. While the first molecular phylogenetic study that included a loriciferan sequence (18S rRNA) failed to establish the phylogenetic position of Loricifera [23], Sørensen et al. [22] detected a sister group relationship between Loricifera and Nematomorpha based on 18S rRNA and histone-3 sequences, although with low nodal support (posterior probability = 0.83). The latter study also detected a sister group relationship between Priapulida and Kinorhyncha, but not monophyly for Cycloneuralia, which several previous molecular studies that lacked loriciferan sequences had indicated [24–29].
The present study investigated the phylogenetic position of phylum Loricifera within Ecdysozoa using nearly complete 18S and 28S rRNA sequences. Also of interest was the phylogenetic status of the taxa Scalidophora and Cycloneuralia.
Materials and methods
Sampling and DNA sequencing
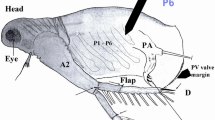
The loriciferan specimen used in this study was collected from Ise Bay, Japan, northwestern Pacific (34°9.77′N, 136°51.40′E, 161–174 m depth) during a cruise of the TR/V Seisui-maru (Mie University) on 21 November 2013. A sediment sample was collected with a biological dredge, subsequently frozen to prevent DNA degradation, and sent to the laboratory. In the laboratory, meiofaunal specimens were extracted by floatation [30] with Ludox® HS 40. The extracted sample was sorted under a stereomicroscope, and a single adult loriciferan specimen (Fig. 1a) was obtained and preserved in 99 % EtOH for DNA extraction.
Total genomic DNA was extracted [31] from the specimen with a DNeasy Tissue Kit (Qiagen, Tokyo). After DNA extraction, the exoskeleton was mounted in Fluoromount G® as a hologenophore (Fig. 1b). The loriciferan specimen was identified as Rugiloricus sp. based on the morphology of the hologenophore.
Nearly complete 18S rRNA (18S) and 28S rRNA (28S) genes sequences were amplified by PCR using previously published primer sets and conditions [31]. All nucleotide sequences were determined by direct sequencing with a BigDye Terminator Kit ver. 3.1 (Life Technologies, Co., USA) and a 3730 DNA Analyzer (Life Technologies, Co., USA). Sequence fragments were assembled by using MEGA 5 [32]. After assembly, 18S (1872 bp) and 28S (3450 bp) sequences were deposited in GenBank under accession numbers LC032019 and LC032020.
Phylogenetic analyses
18S and/or 28S sequences for 66 taxa were obtained from GenBank. We prepared the following five datasets for analyses (Table 1): “18S + 28S (50OTU)” including 18S and 28S sequences for all 50 taxa which both 18S and 28S are available (note that we treated the 18S sequence from Milnesium tardigradum and the 28S sequence from Milnesium sp. as a single OTU, because nearly complete 18S and 28S sequences were unavailable from a single tardigrade species); “18S (50OTU)” including 18S sequences for the same taxa of “18S +28S (50 OTU)”; “28S (50OTU)” including 28S sequences for the same taxa of “18S +28S (50 OTU)”; “18S (65 OTU)” including 18S sequences for more comprehensive taxon sampling especially in Tardigrada, Nematoda, Nematomorpha, Priapulida, and Kinorhyncha than the former three datasets; “18S (63 OTU)” including 18S sequences for same OTU to “18S (65 OTU)” except for Nanaloricus sp. due to its short sequence and Meiopriapulus fijiensis to avoid long branch attraction [22]. Sequences from each gene were pre-aligned separately with MAFFT software [33] using the FFT-NS-2 option and were subsequently divided into domains by eye. Domain sequences were realigned individually with MAFFT software using the L-INS-i option (Additional files 1, 2, 3 and 4). Alignment-ambiguous positions were removed with TrimAl software [34] in “strict setting”, and all positions bearing gaps were also removed. The trimmed domain sequences were recombined to form the final dataset for analysis (Additional files 5, 6, 7 and 8), which was 1426 bp long for 18S and 2189 bp long for 28S in “18S + 28S (50OTU)”, “18S (50OTU)”, and “28S (50OTU), 1277 bp long for 18S in 18S (65 OTU), and 1302 bp long for 18S in 18S (63 OTU). The chi-square test in Kakusan4 [35] indicated that the base composition of each dataset was significantly homogeneous.
Before the analyses, the optimal substitution model was determined with Kakusan4 to be the general time-reversible model with the gamma distribution (GTR + Γ). Phylogenetic trees were constructed by maximum likelihood (ML) implemented in raxmlGUI 1.2 [36, 37], and Bayesian inference (BI) implemented in MrBayes 3.2.1 [38, 39]. Nodal support for the ML tree was assessed through analyses of 1000 bootstrap pseudoreplicates. For BI, Markov-chain Monte-Carlo searches were performed with four chains, each of which was run for 1,000,000 generations, with trees sampled every 100 generations. Stationarity was evaluated by monitoring likelihood values graphically. The initial 20 % of trees from each run were discarded as burn-in, and the remaining trees were used to construct majority-rule consensus trees and determine the Bayesian posterior probability for each clade [39].
Results and discussion
Overall topology in Ecdysozoa
None of the trees conflicted with the others in their overall topology; however, supporting values were lower in datasets with more OTU and shorter sequences (Table 2; Additional files 9, 10, 11 and 12). In our results, increasing the available sequence length with slightly limited taxa generated a better-resolved tree than using more taxa with markedly shortening the sequence length. Thus, we present and mainly discuss the result of 18S + 28S (50 OTU) dataset (Fig. 2). Both the ML and BI trees showed monophyly for the Ecdysozoa (nodal support ML/PP = 99/1.00) as well as for the phyla Priapulida (100/1.00), Nematoda (99/1.00), Nematomorpha (100/1.00), and Arthropoda (89/1.00). Although the monophyly of each phyla were not tested for Kinorhyncha, Loricifera, and Tardigrada in 18S + 28S (50 OTU) dataset, they were supported in 18S (65 OTU) and 18S (63 OTU) with the maximum supporting values (Table 2). Monophyly for Onychophora was not tested due to the inclusion of a single representative of the phylum in all datasets.
Maximum-likelihood tree of 18S + 28S (50 OTU) dataset. The tree is based on 18S + 28S (50 OTU) dataset. Numbers near nodes are the maximum-likelihood bootstrap (BS) and Bayesian posterior probability (PP) values, respectively; values lower than 50 % (BS) or 0.90 (PP) are indicated by dashes. The scale bar indicates branch length in substitutions per site
Within the Ecdysozoa, two basal clades were detected with high nodal support: Priapulida + Kinorhyncha (Scalidophora, excluding Loricifera; nodal support 100/1.00) and Nematoda + Nematomorpha + Loricifera + Tardigrada + Onychophora + Arthropoda (99/1.00). The latter basal clade in turn comprised the clades Nematoda + Nematomorpha clade (= Nematoida), and Loricifera + Tardigrada + Onychophora + Arthropoda clade (= Loricifera + Panarthropoda) in both the ML and BI trees. Support for the Nematoida clade was only moderate (71/0.90), and that for Loricifera + Panarthropoda clade was low (63/0.66). Support for the monophyly of Tardigrada + Onychophora + Arthropoda (= Panarthropoda) was also low (54/0.76). Tardigrada, Onychophora, and Arthropoda formed an unresolved trichotomy.
Phylogenetic evaluation of loricifera, scalidophora, and cycloneuralia
The clade we detected consisting of Loricifera, Nematoida, and Panarthropoda received high nodal support (96/1.00), but the phylogenetic position of Loricifera within this clade remains unclear, as support for the node grouping Loricifera with Panarthropoda was quite low (63/0.66). However, the scalidophoran phyla Priapulida and Kinorhyncha together comprised a clade with high nodal support (100/1.00) to the exclusion of Loricifera, which instead grouped in a highly supported (96/1.00) clade with Nematoida and Panarthropoda. Our results thus do not support both the ‘Scalidophora hypothesis,’ in which Loricifera comprises a clade with Kinorhyncha and Priapulida, and the ‘Loricifera + Nematomorpha hypothesis’. Our trees also indicated non-monophyly for Cycloneuralia, as Loricifera and Nematoida showed closer relationships to Panarthropoda than to other cycloneuralian phyla (Priapulida and Kinorhyncha).
Evaluation of synapomorphies for scalidophora and cycloneuralia
Morphological synapomorphies have previously been proposed that uniting the scalidophoran phyla (Loricifera, Priapulida and Kinorhyncha) and the cycloneuralian phyla (Scalidophora plus Nematoda and Nematomorpha). Putative synapomorphies [11] among Loricifera, Priapulida, and Kinorhyncha include (1) an introvert that has short, spinose scalids that are staggered in arrangement and triradiate in cross-section, and that has (2) inner and outer retractor muscles; (3) a compound filter of protonephridia consisting of two or more terminal cells; (4) basally thickened cuspidate spines; and (5) sensory organs (flosculi) with external cuticular micropapillae and a central pore. The most important synapomorphy proposed for cycloneuralians is the collar-shaped circumoral brain consisting of a ring neuropil [20, 21]. Our results failed to support the monophyly of either Scalidophora or Cycloneuralia, and the putative synapomorphies supporting these groups thus need to be reevaluated.
With regard to the monophyly of Loricifera + Nematoida + Panarthropoda that we detected, three possible topologies among these groups (Fig. 3) in turn suggest two possible evolutionary scenarios for the three scalidophoran phyla (Priapulida, Kinorhyncha, Loricifera). If Loricifera is the sister taxon of Panarthropoda (Fig. 3a) or of Nematoida (Fig. 3b), the most parsimonious scenario is that ‘scalidophoran’ characters arose independently in Loricifera and in the common ancestor of Priapulida + Kinorhyncha and represent convergent characters. Alternatively, if Loricifera is basal in the Loricifera + Nematoida + Panarthropoda clade (Fig. 3c), the most parsimonious interpretation is that the common ancestor of Ecdysozoa possessed ‘scalidophoran’ characters, which the common ancestor of Nematoida and Panarthropoda subsequently lost.
Hypotheses of evolutionary transitions in scalidophoran and cycloneuralian morphological characters. These hypotheses are based on the three possible relationships within the Loricifera + Nematoida + Panarthropoda clade. Sc and Cy above branches indicate morphological characters of the ‘Scalidophora’ and ‘Cycloneuralia,’ respectively; ‘ + ’ and ‘–’ indicate the presence and absence of characters; asterisks indicate well-supported nodes
In all three topologies (Fig. 3), the most parsimonious evolutionary scenario for ‘cycloneuralian’ characters is that they originated once in the common ancestor of Ecdysozoa and were lost once in the common ancestor of Panarthropoda. In other words, the ‘cycloneuralian’ characters are plesiomorphic in ecdysozoans.
Conclusions
We reconstructed the phylogeny of ecdysozoan phyla using nearly complete 18S and 28S rRNA gene sequences, and our results suggested a new hypothesis for the phylogenetic position of Loricifera. These results did not support the previously proposed ‘Scalidophora’ or the ‘Loricifera + Nematomorpha’ clades, but detected a ‘Loricifera + Nematoida + Panarthropoda’ clade with rather high nodal support. Cycloneuralia emerged as paraphyletic, with high nodal support. Relationships among phyla in the ‘Loricifera + Nematoida + Panarthropoda’ clade were not well resolved, and phylogenetic analysis using transcriptomic or genomic data will be necessary to reconstruct the relationships within this clade, and to elucidate evolutionary transitions within Ecdysozoa.
Availability of supporting data
The data sets supporting the results of this article are included within the article and its additional files.
References
Kristensen RM. Loricifera, a new phylum with Aschelminthes characters from the meiobenthos. Z Zool Syst Evol. 1983;21:163–80.
Bang-Berthelsen IH, Schmidt-Rhaesa A, Kristensen RM. Loricifera. In: Schmidt-Rhaesa A. Handbook of zoology. Gastrotricha, Cycloneuralia and Gnathifera. Vol. 1: Nematomorpha, Priapulida, Kinorhyncha, Loricifera. Berlin: De Gruyter; 2013. p. 307–28.
Kristensen RM, Neves RC, Gad G. First report of Loricifera from the Indian Ocean: a new Rugiloricus-species represented by a hermaphrodite. Cah Biol. 2013;54:161–71.
Pardos F, Kristensen RM. First record of Loricifera from the Iberian Peninsula, with the description of Rugiloricus manuelae sp. nov., (Loricifera, Pliciloricidae). Helgoland Mar Res. 2013;67:623–38.
Neves RC, Kristensen RM. A new type of loriciferan larva (Shira larva) from the deep sea of Shatsky Rise Pacific Ocean. Org Divers Evol. 2014;14:163–71.
Neves RC, Gambi C, Danovaro R, Kristensen RM. Spinoloricus cinziae (Phylum Loricifera), a new species from a hypersaline anoxic deep basin in the Mediterranean Sea. Syst Biodivers. 2014;12:489–502.
Danovaro R, Dell’Anno A, Pusceddu A, Gambi C, Heiner I, Kristensen RM. The first metazoa living in permanently anoxic conditions. BioMed Central Biology. 2010;8:30.
Heiner I, Kristensen RM. Urnaloricus gadi nov. gen. et nov. sp. (Loricifera, Urnaloricidae nov. fam.), an aberrant Loricifera with a viviparous pedogenetic life cycle. J Morphol. 2008;270:129–53.
Neuhaus B. Ultrastructure of alimentary canal and body cavity, ground pattern, and phylogenetic relationships of the Kinorhyncha. Microfauna Marina. 1994;9:61–156.
Lemburg C. Ultrastructure of sense organs and receptor cells of the neck and lorica of Halicryptus spinulosus larva (Priapulida). Microfauna Marina. 1995;10:7–30.
Neuhaus B, Higgins RP. Ultrastructure, biology, and phylogenetic relationships of Kinorhyncha. Integr Comp Biol. 2002;42:619–32.
Malakhov VV. Cephalorhyncha, a new type of the animal kingdom uniting Priapulida, Kinorhyncha, and Gordiacea, and a system of Aschelminthes worms. Zool Zh. 1980;59:485–99.
Adrianov AV, Malakhov VV. The phylogeny and classification of the phylum Cephalorhyncha. Zoosyst Ross. 1995;3:181–201.
Adrianov AV, Malakhov VV. The phylogeny and classification of the class Kinorhyncha. Zoosyst Ross. 1996;4:23–44.
Adrianov AV, Malakhov VV. Cephalorhyncha of the world oceans. Moscow: KMK Scientific Press; 1999.
Kristensen RM, Higgins RP. Kinorhyncha. In: Harrison FW, Ruppert EE, editors. Microscopic anatomy of invertebrates, vol. 4. New York: Wiley-Liss; 1991. p. 377–404.
Schmidt-Rhaesa A, Bartolomaeus T, Lemburg C, Ehlers U, Garey JR. The position of the Arthropoda in the phylogenetic system. J Morphol. 1998;238:263–85.
Nielsen C. Animal evolution. 2nd ed. Oxford: Oxford University Press; 2001.
Kristensen RM. Comparative morphology: do the ultrastructural investigations of Loricifera and Tardigrada support the clade Ecdysozoa? In: Legakis A, Sfenthourakis S, Polymeni R, Thessalou-Leagaki M, editors. The new panorama of animal evolution. Proceedings of the 18th International Congress of Zoology. Moscow: Pensoft Publishers, Sofia; 2003. p. 467–77.
Richter S, Loesel R, Purschke G, Schmidt-Rhaesa A, Scholtz G, Stach T, et al. Invertebrate neurophylogeny: suggested terms and definitions for a neuroanatomical glossary. Front Zool. 2010;7:29.
Schmidt-Rhaesa A, Rothe BH, Wägele JW, Bartolomaeus T. Brains in Gastrotricha and Cycloneuralia — a comparison. In: . Deep metazoan phylogeny: the backbone of the Tree of Life. New insights from analyses of molecules, morphology, and theory of data analysis. Berlin: De Gruyter; 2014. p. 93–104.
Sørensen MV, Hebsgaard MB, Heiner I, Glenner H, Willerslev E, Kristensen RM. New data from an enigmatic phylum: evidence from molecular sequence data supports a sister-group relationship between Loricifera and Nematomorpha. J Zool Syst Evol Res. 2008;46:231–9.
Park JK, Rho HS, Kristensen RM, Kim W, Giribet G. First molecular data on the phylum Loricifera — an investigation into the phylogeny of Ecdysozoa with emphasis on the positions of Loricifera and Priapulida. Zool Sci. 2006;23:943–54.
Mallatt J, Giribet G. Further use of nearly complete 28S and 18S rRNA genes to classify Ecdysozoa: 37 more arthropods and kinorhynch. Mol Phylogenet Evol. 2006;40:772–94.
Dunn CW, Hejnol A, Matus DW, Pang K, Browne WE, Smith SA, et al. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature. 2008;452:745–9.
Campbell LI, Rota-Stabelli O, Edgecombe GD, Marchioro T, Longhorn SJ, Telford MJ, et al. MicroRNAs and phylogenomics resolve the relationships of Tardigrada and suggest that velvet worms are the sister group of Arthropoda. Proc Natl Acad Sci U S A. 2011;38:15920–4.
Mallatt J, Craig CW, Yoder MJ. Nearly complete rRNA genes from 371 Animalia: updated structure-based alignment and detailed phylogenetic analysis. Mol Phylogenet Evol. 2012;64:603–17.
Borner J, Rehm P, Schill RO, Ebersberger I, Burmester T. A transcriptome approach to ecdysozoan phylogeny. Mol Phylogenet Evol. 2014;80:79–87.
Rota-Stabelli O, Daley AC, Pisani D. Molecular timetrees reveal a cambrian colonization of land and a new scenario for ecdysozoan evolution. Curr Biol. 2013;23:392–8.
Meiobenthology GO. The microscopic motile fauna of aquatic sediments. Berlin: Springer Verlag; 2009.
Yamasaki H, Hiruta SF, Kajihara H. Molecular phylogeny of kinorhynchs. Mol Phylogenet Evol. 2013;67:303–10.
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol. 2011;28:2731–9.
Katoh K, Toh H. Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics. 2010;26:1899–900.
Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. TrimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–3.
Tanabe AS. KAKUSAN: a computer program to automate the selection of a nucleotide substitution model and the configuration of a mixed model on multilocus data. Mol Ecol Notes. 2007;7:962–4.
Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–90.
Silvestro D, Michalak I. raxmlGUI: a graphical front-end for RAxML. Org Divers Evol. 2012;12:335–7.
Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–4.
Huelsenbeck JP, Ronquist F. MrBayes: Bayesian inference of phylogeny. Bioinformatics. 2001;17:754–5.
Acknowledgements
We thank Dr. Taeko Kimura (Mie University) and the captain and crew of TR/V Seisui-maru (Mie University) for assistance in collecting; Dr. Hiroshi Kajihara (Hokkaido University) for making available molecular laboratory facilities; and Dr. Matthew H. Dick (Hokkaido University) for comments and editing. This study was supported in part by a grant to H. Y. from the International Research Hub Project for Climate Change and Coral Reef/Island Dynamics from the University of the Ryukyus, and a Grant for the Cultivation of Young Scientists to H. Y. from the Research Institute of Marine Invertebrates, Japan.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
H.Y. extracted the specimen, did the molecular laboratory work, and analyzed the data. S. F. identified the specimen to genus. H.Y., S. F., and K. M. discussed the results and wrote the manuscript. All authors read and approved the final manuscript.
Additional files
Additional file 1:
Raw 18S sequence alignment for 18S + 28S (50 OTU) and 18S (50 OTU) datasets. Aligned 18S sequences from 50 species (44 ecdysozoan and six outgroup species) before the removal of alignment-ambiguous positions and gaps.
Additional file 2:
Raw 28S sequence alignment for 18S + 28S (50 OTU) and 28S (50 OTU) datasets. Aligned 28S sequences from 50 species (44 ecdysozoan and six outgroup species) before the removal of alignment-ambiguous positions and gaps.
Additional file 3:
Raw 18S sequence alignment for 18S (65 OTU) dataset. Aligned 18S sequences from 65 species (59 ecdysozoan and six outgroup species) before the removal of alignment-ambiguous positions and gaps.
Additional file 4:
Raw 18S sequence alignment for 18S (63 OTU) dataset. Aligned 18S sequences from 63 species (57 ecdysozoan and six outgroup species) before the removal of alignment-ambiguous positions and gaps.
Additional file 5:
Final 18S sequences for 18S + 28S (50 OTU) and 18S (50 OTU) datasets. Aligned 18S sequences of 50 species after the removal of alignment-ambiguous positions and gaps.
Additional file 6:
Final 28S sequences for 18S + 28S (50 OTU) and 18S (50 OTU) datasets. Aligned 28S sequences of 50 species after the removal of alignment-ambiguous positions and gaps.
Additional file 7:
Final 18S sequences for 18S (65 OTU) dataset. Aligned 18S sequences of 65 species after the removal of alignment-ambiguous positions and gaps.
Additional file 8:
Final 18S sequences for 18S (63 OTU) dataset. Aligned 18S sequences of 63 species after the removal of alignment-ambiguous positions and gaps.
Additional file 9:
Maximum-likelihood tree of 18S (50 OTU) dataset. The tree is based on 18S (50 OTU) dataset. Labelling of values is as in Figure 2.
Additional file 10:
Maximum-likelihood tree of 28S (50 OTU) dataset. The tree is based on 28S (50 OTU) dataset. Labelling of values is as in Figure 2.
Additional file 11:
Maximum-likelihood tree of 18S (65 OTU) dataset. The tree is based on 18S (65 OTU) dataset. Labelling of values is as in Figure 2.
Additional file 12:
Maximum-likelihood tree of 18S (63 OTU) dataset. The tree is based on 18S (63 OTU) dataset. Labelling of values is as in Figure 2.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Yamasaki, H., Fujimoto, S. & Miyazaki, K. Phylogenetic position of Loricifera inferred from nearly complete 18S and 28S rRNA gene sequences. Zoological Lett 1, 18 (2015). https://doi.org/10.1186/s40851-015-0017-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40851-015-0017-0