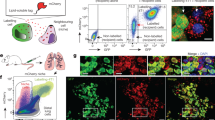
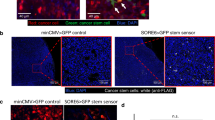
Abstract
Understanding cell–cell interactions is critical in most, if not all, research fields in biology. Nevertheless, studying intercellular crosstalk in vivo remains a relevant challenge, due mainly to the difficulty in spatially locating the surroundings of particular cells in the tissue. Cherry-niche is a powerful new method that enables cells expressing a fluorescent protein to label their surrounding cells, facilitating their specific isolation from the whole tissue as live cells. We previously applied Cherry-niche in cancer research to study the tumor microenvironment (TME) in metastasis. Here we describe how to generate cancer cells with the ability to label their neighboring cells (within the tumor niche) by transferring a liposoluble fluorescent protein. Live niche cells can be isolated and compared with cells distant from the tumor bulk, using a variety of ex vivo approaches. As previously shown, this system has the potential to identify novel components in the TME and improve our understanding of their local interactions. Importantly, Cherry-niche can also be applied to study potential cell–cell interactions due to in vivo proximity in research fields beyond cancer. This protocol takes 2–3 weeks to generate the labeling cells and 1–2 weeks to test their labeling ability.






Similar content being viewed by others
References
Ombrato, L. et al. Metastatic-niche labelling reveals parenchymal cells with stem features. Nature 572, 603–608 (2019).
del Pozo Martin, Y. et al. Mesenchymal cancer cell-stroma crosstalk promotes niche activation, epithelial reversion, and metastatic colonization. Cell Rep. 13, 2456–2469 (2015).
Quail, D. F. & Joyce, J. A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 19, 1423–1437 (2013).
Klemm, F. & Joyce, J. A. Microenvironmental regulation of therapeutic response in cancer. Trends Cell Biol 25, 198–213 (2015).
Barash, S., Wang, W. & Shi, Y. Human secretory signal peptide description by hidden Markov model and generation of a strong artificial signal peptide for secreted protein expression. Biochem. Biophys. Res. Commun. 294, 835–842 (2002).
Flinterman, M. et al. Delivery of therapeutic proteins as secretable TAT fusion products. Mol. Ther. 17, 334–342 (2009).
Shaner, N. C., Steinbach, P. A. & Tsien, R. Y. A guide to choosing fluorescent proteins. Nat. Methods 2, 905–909 (2005).
Espina, V. et al. Laser-capture microdissection. Nat. Protoc. 1, 586–603 (2006).
Legres, L. G., Janin, A., Masselon, C. & Bertheau, P. Beyond laser microdissection technology: follow the yellow brick road for cancer research. Am. J. Cancer Res. 4, 1–28 (2014)
Victora, G. D. et al. Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell 143, 592–605 (2010).
Medaglia, C. et al. Spatial reconstruction of immune niches by combining photoactivatable reporters and scRNA-seq. Science 358, 1622–1626 (2017).
Giladi, A. et al. Dissecting cellular crosstalk by sequencing physically interacting cells. Nat. Biotechnol. 38, 629–637 (2020).
Pasqual, G. et al. Monitoring T cell–dendritic cell interactions in vivo by intercellular enzymatic labelling. Nature 553, 496–500 (2018).
Headley, M. B. et al. Visualization of immediate immune responses to pioneer metastatic cells in the lung. Nature 531, 513–517 (2016).
Calvo, F. et al. Mechanotransduction and YAP-dependent matrix remodelling is required for the generation and maintenance of cancer-associated fibroblasts. Nat. Cell Biol. 15, 637–646 (2013).
Passaro, D. et al. Increased vascular permeability in the bone marrow microenvironment contributes to disease progression and drug response in acute myeloid leukemia. Cancer Cell 32, 324–341.e6 (2017).
Passaro, D., Abarrategi, A., Foster, K., Ariza-McNaughton, L. & Bonnet, D. Bioengineering of humanized bone marrow microenvironments in mouse and their visualization by live imaging. J. Vis. Exp. 2017, 55914 (2017).
Acknowledgements
We thank the Biological Resources Unit, the Flow Cytometry Unit, the Experimental Histopathology Unit and the Cell Services Unit at the Francis Crick Institute for technical support. This work was supported by the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001112), the UK Medical Research Council (FC001112), and the Wellcome Trust (FC001112), and European Research Council grant (ERC CoG-H2020-725492); L.O. was also funded by a Barts Charity Lectureship (grant no. MGU045). D.B. was supported by the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001045), the UK Medical Research Council (FC001045) and the Wellcome Trust (FC001045); D.P. was the recipient of a Junior EHA Fellowship. C.L.C. was supported by Cancer Research UK (CRUK PFA C36195/A26770) and the European Research Council (ERC STG 337066); D.D. was the recipient of an FCT fellowship (SFRH/BD/52195/2013). NESTIN-GFP mice were a kind gift from G. Enikolopov (Stony Brook University).
Author information
Authors and Affiliations
Contributions
L.O. designed the protocol, performed most of the experiments, analyzed the data and wrote the manuscript. E.N. and V.L.B. performed the experiments on liver metastasis and analyzed the data. D.P. and A.W. generated the leukemic labeling ML-1 cells and performed the experiments reported with those cells. I.K. performed the proliferation and the gel contraction experiments and analyzed the data. D.D. and C.L.C. ran pilot experiments to validate the labeling system, which helped with the troubleshooting, and critically read the manuscript. D.B. supervised the experiments with the leukemic cells. I.M. supervised the study and critically revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Protocols thanks Lance Liotta and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key reference using this protocol
Ombrato, L. et al. Nature 572, 603–608 (2019): https://doi.org/10.1038/s41586-019-1487-6
Supplementary information
Supplementary Information
Supplementary Figs. 1–4.
Rights and permissions
About this article
Cite this article
Ombrato, L., Nolan, E., Passaro, D. et al. Generation of neighbor-labeling cells to study intercellular interactions in vivo. Nat Protoc 16, 872–892 (2021). https://doi.org/10.1038/s41596-020-00438-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-020-00438-5
- Springer Nature Limited
This article is cited by
-
Lung endothelium exploits susceptible tumor cell states to instruct metastatic latency
Nature Cancer (2024)
-
Spatial-linked alignment tool (SLAT) for aligning heterogenous slices
Nature Communications (2023)
-
Researchers dig into cancer niches
Nature Methods (2023)
-
Host-derived growth factors drive ERK phosphorylation and MCL1 expression to promote osteosarcoma cell survival during metastatic lung colonization
Cellular Oncology (2023)
-
Secretory GFP reconstitution labeling of neighboring cells interrogates cell–cell interactions in metastatic niches
Nature Communications (2023)