Abstract
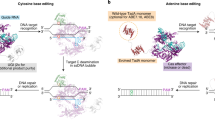
Adenine base editors (ABEs), composed of an engineered deaminase and a catalytically impaired CRISPR–Cas9 variant, are powerful new tools for targeted base editing in cells and organisms. Together with cytosine base editors (CBEs), ABEs enable single-nucleotide conversions cleanly, efficiently and reversibly without double-stranded DNA cleavage, advancing genome editing in a new dimension.

Similar content being viewed by others
References
Kim, H. & Kim, J. S. A guide to genome engineering with programmable nucleases. Nat. Rev. Genet. 15, 321–334 (2014).
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A. & Liu, D. R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016).
Nishida, K. et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 353, aaf8729 (2016).
Zong, Y. et al. Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat. Biotechnol. 35, 438–440 (2017).
Shimatani, Z. et al. Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat. Biotechnol. 35, 441–443 (2017).
Kim, K. et al. Highly efficient RNA-guided base editing in mouse embryos. Nat. Biotechnol. 35, 435–437 (2017).
Wang, L. et al. Enhanced base editing by co-expression of free uracil DNA glycosylase inhibitor. Cell. Res. 27, 1289–1292 (2017).
Komor, A. C. et al. Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C:G-to-T:A base editors with higher efficiency and product purity. Sci. Adv. 3, eaao4774 (2017).
Gaudelli, N. M. et al. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature 551, 464–471 (2017).
Kuscu, C. et al. CRISPR-STOP: gene silencing through base-editing-induced nonsense mutations. Nat. Methods 14, 710–712 (2017).
Billon, P. et al. CRISPR-mediated base editing enables efficient disruption of eukaryotic genes through induction of STOP codons. Mol. Cell. 67, 1068–1079.e4 (2017).
Vierstra, J. et al. Functional footprinting of regulatory DNA. Nat. Methods 12, 927–930 (2015).
Hess, G. T. et al. Directed evolution using dCas9-targeted somatic hypermutation in mammalian cells. Nat. Methods 13, 1036–1042 (2016).
Ma, Y. et al. Targeted AID-mediated mutagenesis (TAM) enables efficient genomic diversification in mammalian cells. Nat. Methods 13, 1029–1035 (2016).
Lee, H. J., Kim, E. & Kim, J. S. Targeted chromosomal deletions in human cells using zinc finger nucleases. Genome Res. 20, 81–89 (2010).
Lee, H. J., Kweon, J., Kim, E., Kim, S. & Kim, J. S. Targeted chromosomal duplications and inversions in the human genome using zinc finger nucleases. Genome Res. 22, 539–548 (2012).
Park, C. Y. et al. Functional correction of large factor VIII gene chromosomal inversions in hemophilia a patient-derived iPSCs using CRISPR-Cas9. Cell. Stem Cell. 17, 213–220 (2015).
Kim, D. et al. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat. Methods 12, 237–243 (2015).
Kim, D. et al. Genome-wide target specificities of CRISPR RNA-guided programmable deaminases. Nat. Biotechnol. 35, 475–480 (2017).
Rees, H. A. et al. Improving the DNA specificity and applicability of base editing through protein engineering and protein delivery. Nat. Commun. 8, 15790 (2017).
Kim, S., Kim, D., Cho, S. W., Kim, J. & Kim, J. S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 24, 1012–1019 (2014).
Woo, J. W. et al. DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat. Biotechnol. 33, 1162–1164 (2015).
Ma, H. et al. Correction of a pathogenic gene mutation in human embryos. Nature 548, 413–419 (2017).
Kleinstiver, B. P. et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529, 490–495 (2016).
Kim, Y. B. et al. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat. Biotechnol. 35, 371–376 (2017).
Acknowledgements
This work was supported by a grant from Institute for Basic Science (IBS-R021-D1).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
J.-S.K. is a co-founder of and holds stock in ToolGen, Inc. focused on genome editing.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kim, JS. Precision genome engineering through adenine and cytosine base editing. Nature Plants 4, 148–151 (2018). https://doi.org/10.1038/s41477-018-0115-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-018-0115-z
- Springer Nature Limited
This article is cited by
-
Recent advances in the plant epitranscriptome
Genome Biology (2023)
-
Split complementation of base editors to minimize off-target edits
Nature Plants (2023)
-
A Review on Advanced CRISPR-Based Genome-Editing Tools: Base Editing and Prime Editing
Molecular Biotechnology (2023)
-
Phytoremediation and sequestration of soil metals using the CRISPR/Cas9 technology to modify plants: a review
Environmental Chemistry Letters (2023)
-
Development of an efficient and precise adenine base editor (ABE) with expanded target range in allotetraploid cotton (Gossypium hirsutum)
BMC Biology (2022)