Abstract
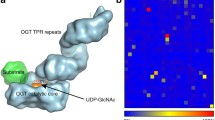
Addition of N-acetylglucosamine (GlcNAc) is a ubiquitous form of intracellular glycosylation catalyzed by the conserved O-linked GlcNAc transferase (OGT). OGT contains an N-terminal domain of tetratricopeptide (TPR) repeats that mediates the recognition of a broad range of target proteins. Components of the nuclear pore complex are major OGT targets, as OGT depletion by RNA interference (RNAi) results in the loss of GlcNAc modification at the nuclear envelope. To gain insight into the mechanism of target recognition, we solved the crystal structure of the homodimeric TPR domain of human OGT, which contains 11.5 TPR repeats. The repeats form an elongated superhelix. The concave surface of the superhelix is lined by absolutely conserved asparagines, in a manner reminiscent of the peptide-binding site of importin α. Based on this structural similarity, we propose that OGT uses an analogous molecular mechanism to recognize its targets.






Similar content being viewed by others
Accession codes
References
Hanover, J.A. Glycan-dependent signaling: O-linked N-acetylglucosamine. FASEB J. 15, 1865–1876 (2001).
Wells, L., Vosseller, K. & Hart, G.W. Glycosylation of nucleocytoplasmic proteins: signal transduction and O-GlcNAc. Science 291, 2376–2378 (2001).
Kearse, K.P. & Hart, G.W. Lymphocyte activation induces rapid changes in nuclear and cytoplasmic glycoproteins. Proc. Natl. Acad. Sci. USA 88, 1701–1705 (1991).
Chou, T.Y., Hart, G.W. & Dang, C.V. c-Myc is glycosylated at threonine 58, a known phosphorylation site and a mutational hot spot in lymphomas. J. Biol. Chem. 270, 18961–18965 (1995).
Slawson, C. & Hart, G.W. Dynamic interplay between O-GlcNAc and O-phosphate: the sweet side of protein regulation. Curr. Opin. Struct. Biol. 13, 631–636 (2003).
Davis, L.I. & Blobel, G. Nuclear pore complex contains a family of glycoproteins that includes p62: glycosylation through a previously unidentified cellular pathway. Proc. Natl. Acad. Sci. USA 84, 7552–7556 (1987).
Hanover, J.A., Cohen, C.K., Willingham, M.C. & Park, M.K. O-linked N-acetylglucosamine is attached to proteins of the nuclear pore. Evidence for cytoplasmic and nucleoplasmic glycoproteins. J. Biol. Chem. 262, 9887–9894 (1987).
Wells, L., Whalen, S.A. & Hart, G.W. O-GlcNAc: a regulatory post-translational modification. Biochem. Biophys. Res. Com. 302, 435–441 (2003).
Yang, X. et al. O-linkage of N-acetylglucosamine to Sp1 activation domain inhibits its transcriptional capability. Proc. Natl. Acad. Sci. USA 98, 6611–6616 (2001).
Zhang, F. et al. O-GlcNAc modification is an endogenous inhibitor of the proteasome. Cell 115, 715–725 (2003).
Kreppel, L.K., Blomberg, M.A. & Hart, G.W. Dynamic glycosylation of nuclear and cytosolic proteins. Cloning and characterization of a unique O-GlcNAc transferase with multiple tetratricopeptide repeats. J. Biol. Chem. 272, 9308–9315 (1997).
Lubas, W.A., Frank, D.W., Krause, M. & Hanover, J.A. O-linked GlcNAc transferase is a conserved nucleocytoplasmic protein containing tetratricopeptide repeats. J. Biol. Chem. 272, 9316–9324 (1997).
Shafi, R. et al. The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc. Natl. Acad. Sci. USA 97, 5735–5739 (2000).
Lubas, W.A. & Hanover, J.A. Functional expression of O-linked GlcNAc transferase. Domain structure and substrate specificity. J. Biol. Chem. 275, 10983–10988 (2000).
Iyer, S.P. & Hart, G.W. Roles of the tetratricopeptide repeat domain in O-GlcNAc transferase targeting and protein substrate specificity. J. Biol. Chem. 278, 24608–24616 (2003).
Kreppel, L.K. & Hart, G.W. Regulation of a cytosolic and nuclear O-GlcNAc transferase. Role of the tetratricopeptide repeats. J. Biol. Chem. 274, 32015–32022 (1999).
Yang, X., Zhang, F. & Kudlow, J.E. Recruitment of O-GlcNAc transferase to promoters by corepressor mSin3A: coupling protein O-GlcNAcylation to transcriptional repression. Cell 110, 69–80 (2002).
D'Andrea, L.D. & Regan, L. TPR proteins: the versatile helix. Trends Biochem. Sci. 28, 655–662 (2003).
Sikorski, R.S., Boguski, M.S., Goebl, M. & Hieter, P. A repeating amino acid motif in CDC23 defines a family of proteins and a new relationship among genes required for mitosis and RNA synthesis. Cell 60, 307–317 (1990).
Das, A.K., Cohen, P.W. & Barford, D. The structure of the tetratricopeptide repeats of protein phosphatase 5: implications for TPR-mediated protein-protein interactions. EMBO J. 17, 1192–1199 (1998).
Gatto, G.J. Jr., Geisbrecht, B.V., Gould, S.J. & Berg, J.M. Peroxisomal targeting signal-1 recognition by the TPR domains of human PEX5. Nat. Struct. Biol. 7, 1091–1095 (2000).
Lapouge, K. et al. Structure of the TPR domain of p67phox in complex with Rac.GTP. Mol. Cell 6, 899–907 (2000).
Scheufler, C. et al. Structure of TPR domain–peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell 101, 199–210 (2000).
Lubas, W.A., Smith, M., Starr, C.M. & Hanover, J.A. Analysis of nuclear pore protein p62 glycosylation. Biochemistry 34, 1686–1694 (1995).
Herold, A., Klymenko, T. & Izaurralde, E. NXF1/p15 heterodimers are essential for mRNA nuclear export in Drosophila. RNA 7, 1768–1780 (2001).
Taylor, P. et al. Two structures of cyclophilin 40: folding and fidelity in the TPR domains. Structure 9, 431–438 (2001).
Kumar, A. et al. An unexpected extended conformation for the third TPR motif of the peroxin PEX5 from Trypanosoma brucei. J. Mol. Biol. 307, 271–282 (2001).
Cingolani, G., Petosa, C., Weis, K. & Muller, C.W. Structure of importin-β bound to the IBB domain of importin-α. Nature 399, 221–229 (1999).
Lee, S.J. et al. The structure of importin-β bound to SREBP-2: nuclear import of a transcription factor. Science 302, 1571–1575 (2003).
Fukuhara, N., Fernandez, E., Ebert, J., Conti, E. & Svergun, D. Conformational variability of nucleo-cytoplasmic transport factors. J. Biol. Chem. 279, 2176–2181 (2004).
Huber, A.H. & Weis, W.I. The structure of the β-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by β-catenin. Cell 105, 391–402 (2001).
Conti, E., Uy, M., Leighton, L., Blobel, G. & Kuriyan, J. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin α. Cell 94, 193–204 (1998).
Young, J.C., Hoogenraad, N.J. & Hartl, F.U. Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell 112, 41–50 (2003).
Fontes, M.R., Teh, T. & Kobe, B. Structural basis of recognition of monopartite and bipartite nuclear localization sequences by mammalian importin-α. J. Mol. Biol. 297, 1183–1194 (2000).
Conti, E. & Kuriyan, J. Crystallographic analysis of the specific yet versatile recognition of distinct nuclear localization signals by karyopherin α. Structure Fold. Des. 8, 329–338 (2000).
Kabsch, W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26, 795–800 (1993).
Terwilliger, T.C. & Berendzen, J. Automated MAD and MIR structure solution. Acta Crystallogr. D 55, 849–861 (1999).
Terwilliger, T.C. Maximum-likelihood density modification. Acta Crystallogr. D 56, 965–972 (2000).
Kleywegt, G.J. & Read, R.J. Not your average density. Structure 5, 1557–1569 (1997).
Jones, T.A., Zou, J.Y., Cowan, S.W. & Kjeldgaard . Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
Brünger, A.T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998).
Brünger, A. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 355, 472–475 (1992).
Acknowledgements
We are grateful to beamline scientists of X06SA at the Swiss Light Source (Zürich) and ESRF ID14-2 (Grenoble) for assistance during data collection. We thank M. Hothorn for introduction to data processing with XDS and P. Brick for critical reading of the manuscript. M.J. was supported by the Human Frontier Science Program Organization (RGP0063/2002-C).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Nucleoporins are major OGT substrates in vivo. (PDF 734 kb)
Rights and permissions
About this article
Cite this article
Jínek, M., Rehwinkel, J., Lazarus, B. et al. The superhelical TPR-repeat domain of O-linked GlcNAc transferase exhibits structural similarities to importin α. Nat Struct Mol Biol 11, 1001–1007 (2004). https://doi.org/10.1038/nsmb833
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb833
- Springer Nature America, Inc.
This article is cited by
-
Motif-dependent binding on the intervening domain regulates O-GlcNAc transferase
Nature Chemical Biology (2023)
-
O-GlcNAcylation: the “stress and nutrition receptor” in cell stress response
Cell Stress and Chaperones (2021)
-
O-GlcNAcylation and its role in the immune system
Journal of Biomedical Science (2020)
-
An intellectual disability syndrome with single-nucleotide variants in O-GlcNAc transferase
European Journal of Human Genetics (2020)
-
O-GlcNAc transferase regulates centriole behavior and intraflagellar transport to promote ciliogenesis
Protein & Cell (2020)