Abstract
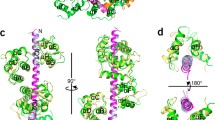
Myosin VI has challenged the lever arm hypothesis of myosin movement because of its ability to take ∼36-nm steps along actin with a canonical lever arm that seems to be too short to allow such large steps. Here we demonstrate that the large step of dimeric myosin VI is primarily made possible by a medial tail in each monomer that forms a rare single α-helix of ∼10 nm, which is anchored to the calmodulin-bound IQ domain by a globular proximal tail. With the medial tail contributing to the ∼36-nm step, rather than dimerizing as previously proposed, we show that the cargo binding domain is the dimerization interface. Furthermore, the cargo binding domain seems to be folded back in the presence of the catalytic head, constituting a potential regulatory mechanism that inhibits dimerization.






Similar content being viewed by others
References
Rock, R.S. et al. Myosin VI is a processive motor with a large step size. Proc. Natl. Acad. Sci. USA 98, 13655–13659 (2001).
Nishikawa, S. et al. Class VI myosin moves processively along actin filaments backward with large steps. Biochem. Biophys. Res. Commun. 290, 311–317 (2002).
Altman, D., Sweeney, H.L. & Spudich, J.A. The mechanism of myosin VI translocation and its load-induced anchoring. Cell 116, 737–749 (2004).
Wells, A.L. et al. Myosin VI is an actin-based motor that moves backwards. Nature 401, 505–508 (1999).
Menetrey, J. et al. The structure of the myosin VI motor reveals the mechanism of directionality reversal. Nature 435, 779–785 (2005).
Bryant, Z., Altman, D. & Spudich, J.A. The power stroke of myosin VI and the basis of reverse directionality. Proc. Natl. Acad. Sci. USA 104, 772–777 (2007).
Park, H. et al. The unique insert at the end of the myosin VI motor is the sole determinant of directionality. Proc. Natl. Acad. Sci. USA 104, 778–783 (2007).
Okten, Z., Churchman, L.S., Rock, R.S. & Spudich, J.A. Myosin VI walks hand-over-hand along actin. Nat. Struct. Mol. Biol. 11, 884–887 (2004).
Yildiz, A. et al. Myosin VI steps via a hand-over-hand mechanism with its lever arm undergoing fluctuations when attached to actin. J. Biol. Chem. 279, 37223–37226 (2004).
Balci, H., Ha, T., Sweeney, H.L. & Selvin, P.R. Interhead distance measurements in myosin VI via SHRImP support a simplified hand-over-hand model. Biophys. J. 89, 413–417 (2005).
Ali, M.Y. et al. Unconstrained steps of myosin VI appear longest among known molecular motors. Biophys. J. 86, 3804–3810 (2004).
Mehta, A.D. et al. Myosin-V is a processive actin-based motor. Nature 400, 590–593 (1999).
Purcell, T.J., Morris, C., Spudich, J.A. & Sweeney, H.L. Role of the lever arm in the processive stepping of myosin V. Proc. Natl. Acad. Sci. USA 99, 14159–14164 (2002).
Veigel, C., Wang, F., Bartoo, M.L., Sellers, J.R. & Molloy, J.E. The gated gait of the processive molecular motor, myosin V. Nat. Cell Biol. 4, 59–65 (2002).
Bahloul, A. et al. The unique insert in myosin VI is a structural calcium-calmodulin binding site. Proc. Natl. Acad. Sci. USA 101, 4787–4792 (2004).
Spudich, J.A. The myosin swinging cross-bridge model. Nat. Rev. Mol. Cell Biol. 2, 387–392 (2001).
Shih, W.M., Gryczynski, Z., Lakowicz, J.R. & Spudich, J.A. A FRET-based sensor reveals large ATP hydrolysis-induced conformational changes and three distinct states of the molecular motor myosin. Cell 102, 683–694 (2000).
Forkey, J.N., Quinlan, M.E., Shaw, M.A., Corrie, J.E. & Goldman, Y.E. Three-dimensional structural dynamics of myosin V by single-molecule fluorescence polarization. Nature 422, 399–404 (2003).
Menetrey, J., Llinas, P., Mukherjea, M., Sweeney, H.L. & Houdusse, A. The structural basis for the large powerstroke of myosin VI. Cell 131, 300–308 (2007).
Rock, R.S. et al. A flexible domain is essential for the large step size and processivity of myosin VI. Mol. Cell 17, 603–609 (2005).
Knight, P.J. et al. The predicted coiled-coil domain of myosin 10 forms a novel elongated domain that lengthens the head. J. Biol. Chem. 280, 34702–34708 (2005).
Lister, I. et al. A monomeric myosin VI with a large working stroke. EMBO J. 23, 1729–1738 (2004).
Altman, D., Goswami, D., Hasson, T., Spudich, J.A. & Mayor, S. Precise positioning of myosin VI on endocytic vesicles in vivo. PLoS Biol. 5, e210 (2007).
Spudich, G. et al. Myosin VI targeting to clathrin-coated structures and dimerization is mediated by binding to Disabled-2 and PtdIns(4,5)P2. Nat. Cell Biol. 9, 176–183 (2007).
Park, H. et al. Full-length myosin VI dimerizes and moves processively along actin filaments upon monomer clustering. Mol. Cell 21, 331–336 (2006).
Berger, B. et al. Predicting coiled coils by use of pairwise residue correlations. Proc. Natl. Acad. Sci. USA 92, 8259–8263 (1995).
Uversky, V.N. Natively unfolded proteins: a point where biology waits for physics. Protein Sci. 11, 739–756 (2002).
Bonneau, R. et al. De novo prediction of three-dimensional structures for major protein families. J. Mol. Biol. 322, 65–78 (2002).
Marqusee, S. & Baldwin, R.L. Helix stabilization by Glu-...Lys+ salt bridges in short peptides of de novo design. Proc. Natl. Acad. Sci. USA 84, 8898–8902 (1987).
O'Shea, E.K., Rutkowski, R. & Kim, P.S. Evidence that the leucine zipper is a coiled coil. Science 243, 538–542 (1989).
Zaman, M.H., Berry, R.S. & Sosnick, T.R. Entropic benefit of a cross-link in protein association. Proteins 48, 341–351 (2002).
Rock, R.S., Rief, M., Mehta, A.D. & Spudich, J.A. In vitro assays of processive myosin motors. Methods 22, 373–381 (2000).
Lin, H.P. et al. Cell adhesion molecule Echinoid associates with unconventional myosin VI/Jaguar motor to regulate cell morphology during dorsal closure in Drosophila. Dev. Biol. 311, 423–433 (2007).
Wang, E. & Wang, C.L. (i, i + 4) Ion pairs stabilize helical peptides derived from smooth muscle caldesmon. Arch. Biochem. Biophys. 329, 156–162 (1996).
Kuhlman, B., Yang, H.Y., Boice, J.A., Fairman, R. & Raleigh, D.P. An exceptionally stable helix from the ribosomal protein L9: implications for protein folding and stability. J. Mol. Biol. 270, 640–647 (1997).
Howard, J. Mechanics of Motor Proteins and the Cytoskeleton Vol. xvi 367 (Sinauer Associates, Sunderland, 2001).
Idiris, A., Alam, M.T. & Ikai, A. Spring mechanics of α-helical polypeptide. Protein Eng. 13, 763–770 (2000).
Zagrovic, B., Jayachandran, G., Millett, I.S., Doniach, S. & Pande, V.S. How large is an α-helix? Studies of the radii of gyration of helical peptides by small-angle X-ray scattering and molecular dynamics. J. Mol. Biol. 353, 232–241 (2005).
Sun, Y. et al. Myosin VI walks “wiggly” on actin with large and variable tilting. Mol. Cell 28, 954–964 (2007).
De La Cruz, E.M., Ostap, E.M. & Sweeney, H.L. Kinetic mechanism and regulation of myosin VI. J. Biol. Chem. 276, 32373–32381 (2001).
Chen, Y.H., Yang, J.T. & Chau, K.H. Determination of the helix and β form of proteins in aqueous solution by circular dichroism. Biochemistry 13, 3350–3359 (1974).
Lipfert, J., Millett, I.S., Seifert, S. & Doniach, S. Sample holder for small-angle X-ray scattering static and flow cell measurements. Rev. Sci. Instrum. 77, 046108 (2006).
Beno, M. et al. Basic energy sciences synchrotron radiation center undulator sector at the advanced photon source. Nucl. Instrum. Methods Phys. Res. A 467–468, 690–693 (2001).
Seifert, S., Winans, R.E., Tiede, D.M. & Thiyagarajan, P. Design and performance of a ASAXS instrument at the Advanced Photon Source. J. Appl. Cryst. 33, 782–784 (2000).
Guinier, A. La diffraction des rayons X aux tres petits angles: Application á l'etude de phenomenes ultramicroscopiques. Ann. Phys. (Paris) 12, 161–237 (1939).
Svergun, D. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 25, 495–503 (1992).
Svergun, D.I. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 76, 2879–2886 (1999).
Svergun, D.I., Petoukhov, M.V. & Koch, M.H. Determination of domain structure of proteins from X-ray solution scattering. Biophys. J. 80, 2946–2953 (2001).
Kozin, M.B. & Svergun, D.I. Automated matching of high- and low-resolution structural models. J. Appl. Crystallogr. 34, 33–41 (2001).
Volkov, V.V. & Svergun, D.I. Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 36, 860–864 (2003).
Wriggers, W. & Chacon, P. Using Situs for the registration of protein structures with low-resolution bead models from X-ray solution scattering. J. Appl. Crystallogr. 34, 773–776 (2001).
Churchman, L.S., Okten, Z., Rock, R.S., Dawson, J.F. & Spudich, J.A. Single molecule high-resolution colocalization of Cy3 and Cy5 attached to macromolecules measures intramolecular distances through time. Proc. Natl. Acad. Sci. USA 102, 1419–1423 (2005).
Rice, S.E., Purcell, T.J. & Spudich, J.A. Building and using optical traps to study properties of molecular motors. Methods Enzymol. 361, 112–133 (2003).
Holmes, K.C., Angert, I., Kull, F.J., Jahn, W. & Schroder, R.R. Electron cryo-microscopy shows how strong binding of myosin to actin releases nucleotide. Nature 425, 423–427 (2003).
Pettersen, E.F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Acknowledgements
We thank A. Dunn, Z. Bryant and N. Geething of Stanford University for technical help with protein purification and motility assays, critical discussions and manuscript review; H.L. Sweeney of the University of Pennsylvania for plasmids; K. Holmes of the Max Planck Institute for Medical Research, Heidelberg, for the acto-myosin PDB model; T. Fenn of Stanford University for technical help with MALS analysis and graphical presentation; T. Purcell of the University of California, San Fransisco, for help with model building; S. Seifert of the Advanced Photon Source; R. Fenn of Stanford University for help with SAXS data collection; and S. Patel of Stanford University for MALDI analysis. Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. B.J.S. is partially supported by grant T32 GM008294; S.D. is supported by grant PO1 GM066275; and J.A.S. is supported by grant GM33289, all from the US National Institutes of Health.
Author information
Authors and Affiliations
Contributions
B.J.S. designed constructs, purified proteins, collected CD, MALS, DLS, gel filtration, motility and trap data, and built models; S.S. purified proteins, collected motility and trap data, and built models; J.L. collected and analyzed SAXS data; S.D. and J.A.S. helped in study design and data interpretation; all authors discussed the results and offered revisions on the manuscript.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 and Supplementary Methods (PDF 604 kb)
Rights and permissions
About this article
Cite this article
Spink, B., Sivaramakrishnan, S., Lipfert, J. et al. Long single α-helical tail domains bridge the gap between structure and function of myosin VI. Nat Struct Mol Biol 15, 591–597 (2008). https://doi.org/10.1038/nsmb.1429
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1429
- Springer Nature America, Inc.
This article is cited by
-
How myosin VI traps its off-state, is activated and dimerizes
Nature Communications (2023)
-
Characterization of long and stable de novo single alpha-helix domains provides novel insight into their stability
Scientific Reports (2017)
-
NDP52 activates nuclear myosin VI to enhance RNA polymerase II transcription
Nature Communications (2017)
-
The myosin X motor is optimized for movement on actin bundles
Nature Communications (2016)
-
A programmable DNA origami nanospring that reveals force-induced adjacent binding of myosin VI heads
Nature Communications (2016)