Abstract
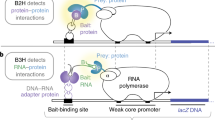
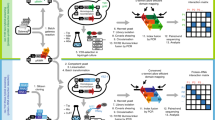
Biochemical 'pathways' are systems of dynamically assembling and disassembling protein complexes, and thus, much of modern biological research is concerned with how, when and where proteins interact with other proteins involved in biochemical processes. The demand for simple approaches to study protein–protein interactions, particularly on a large scale, has grown recently with the progress in genome projects, as the association of unknown with known gene products provides one crucial way of establishing the function of a gene. It was with this challenge in mind that our laboratory developed a simple survival protein-fragment complementation assay (PCA) based on the enzyme dihydrofolate reductase (DHFR). In the DHFR PCA strategy, two proteins of interest are fused to complementary fragments of DHFR. If the proteins of interest interact physically, the DHFR complementary fragments are brought together and fold into the native structure of the enzyme, reconstituting its activity, detectable by the survival of cells expressing the fusion proteins and growth in selective medium. Using the protocol described here, the survival selection can be completed in one to several days, depending on the cell type.



Similar content being viewed by others
References
Michnick, S.W. Exploring protein interactions by interaction-induced folding of proteins from complementary peptide fragments. Curr. Opin. Struct. Biol. 11, 472–477 (2001).
Campbell-Valois, F.X. & Michnick, S. Chemical biology on PINs and NeeDLes. Curr. Opin. Chem. Biol. 9, 31–37 (2005).
Remy, I. & Michnick, S.W. Application of protein-fragment complementation assays in cell biology. Biotechniques 42, 137–145 (2007).
Remy, I., Campbell-Valois, F.X., Ghaddar, G., Aquin, S. & Michnick, S.W. Detection of protein interactions and library screening with protein-fragment complementation assays. In Protein–Protein Interactions: A Molecular Cloning Manual Ch. 33, 2nd edn. (eds. Golemis, E.A. & Adams, P.D.), 637–672 (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, 2005).
Remy, I. & Michnick, S.W. Clonal selection and in vivo quantitation of protein interactions with protein-fragment complementation assays. Proc. Natl. Acad. Sci. USA 96, 5394–5399 (1999).
Pelletier, J.N., Campbell-Valois, F.X. & Michnick, S.W. Oligomerization domain-directed reassembly of active dihydrofolate reductase from rationally designed fragments. Proc. Natl. Acad. Sci. USA 95, 12141–12146 (1998).
Remy, I. & Michnick, S.W. A cDNA library functional screening strategy based on fluorescent protein complementation assays to identify novel components of signaling pathways. Methods 32, 381–388 (2004).
Galarneau, A., Primeau, M., Trudeau, L.E. & Michnick, S.W. Beta-lactamase protein fragment complementation assays as in vivo and in vitro sensors of protein protein interactions. Nat. Biotechnol. 20, 619–622 (2002).
Wehrman, T., Kleaveland, B., Her, J.H., Balint, R.F. & Blau, H.M. Protein–protein interactions monitored in mammalian cells via complementation of beta-lactamase enzyme fragments. Proc. Natl. Acad. Sci. USA 99, 3469–3474 (2002).
Luker, K.E. et al. Kinetics of regulated protein–protein interactions revealed with firefly luciferase complementation imaging in cells and living animals. Proc. Natl. Acad. Sci. USA 101, 12288–12293 (2004).
Paulmurugan, R. & Gambhir, S.S. Monitoring protein–protein interactions using split synthetic Renilla luciferase protein-fragment-assisted complementation. Anal. Chem. 75, 1584–1589 (2003).
Paulmurugan, R., Umezawa, Y. & Gambhir, S.S. Noninvasive imaging of protein–protein interactions in living subjects by using reporter protein complementation and reconstitution strategies. Proc. Natl. Acad. Sci. USA 99, 15608–15613 (2002).
Ghosh, I., Hamilton, A.D. & Regan, L. Antiparallel leucine zipper-directed protein reassembly: application to the green fluorescent protein. J. Am. Chem. Soc. 122, 5658–5659 (2000).
Hu, C.D., Chinenov, Y. & Kerppola, T.K. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 9, 789–798 (2002).
Remy, I., Montmarquette, A. & Michnick, S.W. PKB/Akt modulates TGF-beta signalling through a direct interaction with Smad3. Nat. Cell Biol. 6, 358–365 (2004).
Remy, I. & Michnick, S.W. A highly sensitive protein–protein interaction assay based on Gaussia luciferase. Nat. Methods 3, 977–979 (2006).
Vidal, M. & Legrain, P. Yeast forward and reverse 'n'-hybrid systems. Nucleic Acids Res. 27, 919–929 (1999).
Walhout, A.J. et al. Protein interaction mapping in C. elegans using proteins involved in vulval development. Science 287, 116–122 (2000).
Uetz, P. et al. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature 403, 623–627 (2000).
Rain, J.C. et al. The protein–protein interaction map of Helicobacter pylori. Nature 409, 211–215 (2001).
Israel, D.I. & Kaufman, R.J. Dexamethasone negatively regulates the activity of a chimeric dihydrofolate reductase/glucocorticoid receptor protein. Proc. Natl. Acad. Sci. USA 90, 4290–4294 (1993).
Thillet, J., Absil, J., Stone, S.R. & Pictet, R. Site-directed mutagenesis of mouse dihydrofolate reductase. Mutants with increased resistance to methotrexate and trimethoprim. J. Biol. Chem. 263, 12500–12508 (1988).
Remy, I. & Michnick, S.W. Visualization of biochemical networks in living cells. Proc. Natl. Acad. Sci. USA 98, 7678–7683 (2001).
Campbell-Valois, F.X., Tarassov, K. & Michnick, S.W. Massive sequence perturbation of a small protein. Proc. Natl. Acad. Sci. USA 102, 14988–14993 (2005).
Pelletier, J.N., Arndt, K.M., Pluckthun, A. & Michnick, S.W. An in vivo library-versus-library selection of optimized protein–protein interactions. Nat. Biotechnol. 17, 683–690 (1999).
Campbell-Valois, F.X. & Michnick, S.W. Synthesis of Degenerated Libraries of the RBD of Raf and Rapid Selection of Fast-folding and Stable Clones with the DHFR Protein Fragment Complementation Assay (Humana Press, New Jersey, 2006).
Kaufman, R.J. Selection and coamplification of heterologous genes in mammalian cells. Methods Enzymol. 185, 537–566 (1990).
Acknowledgements
This work was supported by the Canadian Institute of Health Research.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Remy, I., Campbell-Valois, F. & Michnick, S. Detection of protein–protein interactions using a simple survival protein-fragment complementation assay based on the enzyme dihydrofolate reductase. Nat Protoc 2, 2120–2125 (2007). https://doi.org/10.1038/nprot.2007.266
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2007.266
- Springer Nature Limited
This article is cited by
-
Expression, Purification and Crystallization of Asrij, A Novel Scaffold Transmembrane Protein
The Journal of Membrane Biology (2021)
-
Disease-causing mutations in subunits of OXPHOS complex I affect certain physical interactions
Scientific Reports (2019)
-
Dissecting the dimerization motif of Enterococcus hirae’s Zn(II)CopY
JBIC Journal of Biological Inorganic Chemistry (2012)
-
Using the β-lactamase protein-fragment complementation assay to probe dynamic protein–protein interactions
Nature Protocols (2007)