Abstract
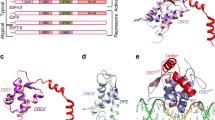
SAP-1 and Elk-1 are members of a large group of eukaryotic transcription factors that contain a conserved ETS DNA binding domain and that cooperate with the serum response factor (SRF) to activate transcription of the c-fos protooncogene. Despite the high degree of sequence similarity, which includes an identical amino acid sequence for the DNA recognition helix within the ETS domain of these proteins, they exhibit different DNA binding properties. Here we report the 2.1 Å crystal structure of the ETS domain of Elk-1 bound to a high affinity E74 DNA (E74DNA) site and compare it to a SAP-1–E74DNA complex. This comparison reveals that the differential DNA binding properties of these proteins are mediated by non-conserved residues distal to the DNA binding surface that function to orient conserved residues in the DNA recognition helix for protein-specific DNA contacts. As a result, nearly one-third of the interactions between the protein recognition helix and the DNA are different between the SAP-1 and Elk-1 DNA complexes. Taken together, these studies reveal a novel mechanism for the modulation of DNA binding specificity within a conserved DNA binding domain, and have implications for how highly homologous ETS proteins exhibit differential DNA-binding properties.




Similar content being viewed by others
Accession codes
References
Sharrocks, A.D., Brown, A.L., Ling, Y. & Yates, P.R. Int. J. of Biochem. Cell Biol. 29, 1371–1387 (1997).
Dittmer, J. & Nordheim, A. Biochim. Biophys. Acta Rev. Cancer 1377, F1–F11 (1998).
Graves, B.J. & Petersen, J.M. In Advances in Cancer Research (Woude, G.v.d. & Klein, G., eds., Academic Press, San Diego), 1–55 (1998).
Yang, S.H., Yates, P.R., Mo, Y. & Sharrocks, A.D. Gene Ther. Mol. Biol. 3, 355–371 (1998).
Shore, P. & Sharrocks, A.D. Eur. J. Biochem. 229, 1–13 (1995).
Shore, P. & Sharrocks, A.D. Nucleic Acids Res. 23, 4698–4706 (1995).
Shore, P. et al. Mol. Cell. Biol. 16, 3338–3349 (1996).
Mo, Y., Vaessen, B., Johnston, K. & Marmorstein, R. Mol. Cell 2, 201–212 (1998).
Brennan, R.G. Cell 74, 773–776 (1993).
Werner, M.H. et al. J. Biomol. NMR 10, 317–328 (1997).
Kodandapani, R. et al. Nature 380, 456–460 (1996).
Batchelor, A.H., Piper, D.E., de la Brousse, F.C., Mcknight, S.L. & Wolberger, C. Science 279, 1037–1041 (1998).
Donaldson, L.W., Petersen, J.M., Graves, B.J. & McIntosh, L.P. EMBO J. 15, 125–134 (1996).
Aggarwal, A.K. Methods Enzymol . 1, 83–90 (1990).
Otwinowski, Z. In Proceedings of the CCP4 Study Weekend: Data collection and Processing. (Sawyer, L., Isaacs, N. & Bailey, S., eds., SERC Daresbury Laboratory, Warrington, UK), 56–62 (1993).
Navaza, J. Acta Crystallogr. A 50, 157–163 (1994).
Brunger, A.T. X-PLOR 3.1: a system for X-ray crystallography and NMR. (Yale University Press, New Haven, Connecticut; 1992).
Brunger, A.T. Nature 335, 472–474 (1992).
Jones, T.A., Zou, J.Y. & Cowen, S.W. Acta Crystallogr. A 47, 110–119 (1991).
Brunger, A.T. & Krukowski, A. Acta Crystallogr. A 46, 585–593 (1990).
Rice, L.M. & Brunger, A.T. Proteins 19, 277–290 (1994).
Kleywegt, G.J. & Jones, T.A. Acta Crystallogr. D 52, 829–832 (1996).
Laskowski, R.A., MacArthur, M.A., Moss, D.S. & Thornton, J.M. J. Appl. Crystallogr. 26, 283–291 (1993).
Jiang, J.S. & Brunger, A.T. J. Mol. Biol. 243, 100–115 (1994).
Brunger, A.T. et al. Acta Crystallogr. D 54, 905–921 (1998).
Hutchinson, E.G. & Thornton, J.M. Protein Sci. 5, 212–220 (1996).
Acknowledgements
We thank R. Treisman (Imperial Cancer Research Fund) for providing the pElk-1 plasmid, and R. Venkataramani, X. Li and R. Trievel for collecting data at NSLS and useful discussions. This work was supported by a grant from NIH and ACS to R.M.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mo, Y., Vaessen, B., Johnston, K. et al. Structure of the Elk-1–DNA complex reveals how DNA-distal residues affect ETS domain recognition of DNA. Nat Struct Mol Biol 7, 292–297 (2000). https://doi.org/10.1038/74055
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/74055
- Springer Nature America, Inc.
This article is cited by
-
Cyclin-dependent kinase 2 (CDK2) is a key mediator for EGF-induced cell transformation mediated through the ELK4/c-Fos signaling pathway
Oncogene (2016)
-
An affinity-structure database of helix-turn-helix: DNA complexes with a universal coordinate system
BMC Bioinformatics (2015)
-
DNA-dependent formation of transcription factor pairs alters their binding specificity
Nature (2015)
-
Iron deficiency upregulates Egr1 expression
Genes & Nutrition (2015)
-
Monomeric and Dimeric Models of ERK2 in Conjunction with Studies on Cellular Localization, Nuclear Translocation, and In Vitro Analysis
Molecules and Cells (2012)