Abstract
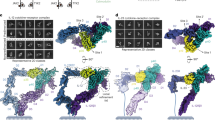
Interleukin-1 (IL-1) is an important mediator of inflammatory disease. The IL-1 family currently consists of two agonists, IL-1α and IL-lβ, and one antagonist, IL-1ra. Each of these molecules binds to the type I IL-1 receptor (IL1R)1. The binding of IL-1α or IL-1β to IL1R is an early step in IL-1 signal transduction and blocking this interaction may therefore be a useful target for the development of new drugs. Here we report the three-dimensional structure of IL-1β bound to the extracellular domain of IL1R (s-IL1R) at 2.5Å resolution. IL-1β binds to s-IL1R with a 1:1 stoichiometry. The crystal structure shows that s-IL1R consists of three immunoglobulin-like domains which wrap around IL-1β in a manner distinct from the structures of previously described cytokine–receptor complexes. The two receptor-binding regions on IL-1β identified by site-directed mutagenesis2,3 both contact the receptor: one binds to the first two domains of the receptor, while the other binds exclusively to the third domain.
Similar content being viewed by others
References
Dinarello, C. A. Biologic basis for interleukin-1 in disease. Blood 87, 2095–2147 (1996).
Labriola-Thompkins, E. et al. Identification of the discontinuous binding site in human interleukin 1β for the type I interleukin 1 receptor. Proc. Natl Acad. Sci. USA 88, 11182–11186 (1991).
Evans, R. J. et al. Mapping receptor binding sites in interleukin (IL)-l receptor antagonist and IL-1β by site-directed mutagenesis. J. Biol. Chem. 270, 11477–11483 (1995).
Sims, J. E. et al. Cloning the interleukin 1 receptor from human T cells. Proc. Natl Acad. Sci. USA 86, 8946–8950 (1989).
Vigers, G. P. A. et al. X-ray structure of interleukin-1 receptor antagonist at 2.0 Å resolution. J. Biol. Chem. 269, 12874–12879 (1994).
Bork, P., Holm, L. & Sander, C. The immunoglobulin fold. Structural classification, sequence patterns and common core. J. Mol. Biol. 242, 309–320 (1994).
Shapiro, L. et al. Structural basis of cell-cell adhesion by cadherins. Nature 374, 327–337 (1995).
Priestle, J. P., Schaer, H. P. & Gruetter, M. G. Crystallographic refinement of interleukin 1 beta at 2.0 Å resolution. Proc. Natl Acad. Sci. USA 86, 9667–9671 (1989).
Walter, M. R. et al. Crystal structure of a complex between interferon-γ and its soluble high-affinity receptor. Nature 376, 230–235 (1995).
de Vos, A. M., Ultsch, M. & Kossiakoff, A. A. Human growth hormone and extracellular domain of its receptor: crystal structure of the complex. Science 255, 306–312 (1992).
Livnah, O. et al. Functional mimicry of a protein hormone by a peptide agonist: the EPO receptor complex at 2.8 Å. Science 273, 464–471 (1996).
Banner, D. W. et al. Crystal structure of the soluble human 55 kd TNF receptor-human TNF beta complex: implications for TNF receptor activation. Cell 73, 431–445 (1993).
Greenfeder, S. A. et al. Molecular cloning and characterization of a second subunit of the interleukin 1 receptor complex. J. Biol. Chem. 270, 1357–13765 (1995).
Otwinowski, Z. in Data Collection and Processing (eds Sawyer, L., Isaacs, N. & Bailey, S.) (SERC Daresbury Laboratory, Warrington, UK, 1993).
CCP4: Collaborative Computational Project No 4, Daresbury UK. Acta Crystallogr. D 50, 760 (1994).
Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
Brunger, A. T., Kuriyan, J. & Karplus, M. Crystallographic R factor refinement by molecular dynamics. Science 235, 458–460 (1987).
Lee, B. & Richards, F. M. The interpretation of protein structures: estimation of static accessibility. J. Mol. Biol. 55, 379–400 (1971).
Evans, S. V. SETOR: hardware-lighted three-dimensional solid model representations of macromolecules. J. Mol. Graphics 11, 134–138 (1993).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Vigers, G., Anderson, L., Caffes, P. et al. Crystal structure of the type-I interleukin-1 receptor complexed with interleukin-1β. Nature 386, 190–194 (1997). https://doi.org/10.1038/386190a0
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/386190a0
- Springer Nature Limited
This article is cited by
-
Discovery of a selective and biologically active low-molecular weight antagonist of human interleukin-1β
Nature Communications (2023)
-
Optimization of IL-1RA structure to achieve a smaller protein with a higher affinity to its receptor
Scientific Reports (2022)
-
Evaluation of wound healing effect of Mallotus philippensis (Lam.) Mull. Arg. by in silico multitargets directed for multiligand approach
In Silico Pharmacology (2022)
-
Rifampicin and Letermovir as potential repurposed drug candidate for COVID-19 treatment: insights from an in-silico study
Pharmacological Reports (2021)
-
Comparative Analyses of the Conformational Dynamics Between the Soluble and Membrane-Bound Cytokine Receptors
Scientific Reports (2020)