Abstract
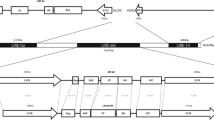
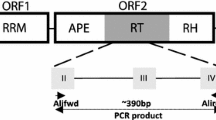
We have isolated and characterized conserved regions of the reverse transcriptase gene from non-LTR retrotransposons, also called long interspersed nuclear elements (LINEs), from Beta vulgaris, B. lomatogona and B. nana. The novel elements show strong homology to other non-LTR retrotransposons from plants, man and animals. LINEs are present in all species of the genus Beta tested, but there was variation in copy number. Analysis by Southern hybridization and fluorescent in situ hybridization revealed the clustered organization of these retroelements in beet species. PCR amplification using degenerate primers to conserved motifs of the predicted LINE protein sequence enabled the cloning of LINEs from both Monocotyledonae (Allium cepa, Oryza sativa and Secale cereale) and Dicotyledonae (Nicotiana tabacum and Antirrhinum majus) indicating that LINEs are a universal feature of plant genomes. A dendrogram of fifteen new and six previously isolated sequences showed the high level of sequence divergence while revealing families characteristic of some genera. The genomic organization of non-LTR retrotransposons was examined more detailed in A. majus and O. sativa.
Similar content being viewed by others
References
Bennetzen JL: The contribution of retroelements to plant genome organization, function and evolution. Trends Microbiol 4: 347–353 (1996).
Boeke JD: LINEs and Alus: the polyA connection. Nature Genet 16: 6–7 (1997).
Brandes A, HeslopHarrison JS, Kamm A, Kubis S, Doudrick RL, Schmidt T: Comparative analysis of the chromosomal and genomic organization of Ty1copialike retrotransposons in pteridophytes, gymnosperms and angiosperms. Plant Mol Biol 33: 11–21 (1997).
Burke WD, Calalang CC, Eickbush TH: The sitespecific ribosomal insertion element type II of Bombyx mori (R2Bm) contains the coding sequence for a reverse transcriptaselike enzyme. Mol Cell Biol 7: 2221–2230 (1987).
Dombroski BA, Mathias SL, Nanthakumar E, Scott AF, Kazazian HH Jr: Isolation of an active human transposable element. Science 254: 1805–1808 (1991).
Eickbush TH: Origin and Evolutionary Relationships of Retroelements. In: Morse SS (ed) The Evolutionary Biology of Viruses, pp. 121–157. Raven Press, New York (1994).
Fawcett DH, Lister CK, Kellett E, Finnegan DJ: Transposable elements controlling IR hybrid dysgenesis in D. melanogaster are similar to mammalian LINEs. Cell 47: 1007–1015 (1986).
Feinberg AP, Vogelstein B: A technique for radiolabelling DNArestriction endonuclease fragments to high specific activity. Ann Biochem 137: 266–267 (1983).
Flavell AJ, Smith DB, Kumar A: Extreme heterogenity of Ty1copia group retrotransposons in plants. Mol Gen Genet 231: 233–242 (1992).
Flavell AJ, Dunbar E, Anderson R, Pearce SR, Hartley R, Kumar A: Ty1copia group retrotransposons are ubiquitous and heterogeneous in higher plants. Nucl Acids Res 20: 3639–3644 (1992).
Garrett JE, Knutzon DS, Carroll D: Composite transposable elements in the Xenopus laevis genome. Mol CellBiol 9: 3018–3027 (1989).
HeslopHarrison JS, Brandes A, Taketa S, Schmidt T, Vershinin AV, Alkhimova EG, Kamm A, Doudrick RL, Schwarzacher T, Katsiotis A, Kubis S, Kumar A, Pearce SR, Flavell AJ, Harrison GE: The chromosomal distributions of Ty1copia group retrotransposable elements in higher plants and their implications for genome evolution. Genetica, in press.
Higashiyama T, Noutoshi Y, Fujie M, Yamada T:Zepp, aLINElike retrotransposon accumulated in the Chlorella telomeric region. EMBO J 16: 3715–3723 (1997).
Hirochika H, Hirochika R: Ty1copia group retrotransposons as ubiquitous components of plant genomes. Jpn J Genet 68: 35–46 (1993).
Hutchinson CA, Hardies SC, Loeb DD, Shehee WR, Edgell MH: LINEs and related retrotransposons: long interspersed repeated sequences in the eukaryotic genome. In: Berg DE, Howe MM(eds)Mobile DNA, pp. 593–617. American Society for Microbiology, Washington, DC (1989).
Kimmel BE, ole-Moiyoi OK, Young JR: Ingi, a 5.2kb dispersed sequence element from Trypanosoma brucei that carries half of a smaller mobile element at either end and has homology to mammalian LINEs. Mol Cell Biol 7: 1465–1475 (1987).
Kohlstaedt LA, Wang J, Friedmann JM, Rice PA, Steitz TA: Crystal structure at 3.5 Å resolution of HIV1 reverse transcriptase complexed with an inhibitor. Science 256: 1783–1790 (1992).
Kubis S, HeslopHarrison JS, Schmidt T: A family of differentially amplified repetitive DNA sequences in the genus Beta reveals genetic variation in Beta vulgaris subspecies and cultivars. J Mol Evol 44: 310–320 (1997).
Kumar S, Tamura K, Nei M: MEGA: Molecular Evolutionary Genetics Analysis, version 1.0. Pennsylvania State University, University Park, PA 16802 (1993).
Larder BA, Kemp SD, Purifoy DJM: Infectious potential of human immunodeficiency virus type I reverse transcriptase mutants with altered inhibitor sensitivity. Proc Natl Acad Sci USA 86: 4803 (1989).
Leeton PRJ, Smyth DR: An abundant LINElike element amplified in the genome of Lilium speciosum. Mol Gen Genet 237: 97–104 (1993).
Lindauer A, Fraser D, Brüderlein M, Schmitt R: Reverse transcriptase families and a copialike retrotransposon, Osser, in the green alga Volvox carteri. FEBS Lett 319: 261–266 (1993).
Luan DD, Korman MH, Jakubczak JL, Eickbush TH: Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: amechanism for nonLTR retrotransposition. Cell 72: 595–605 (1993).
Marschalek R, Hofmann J, Schumann G, Gösseringer R, Dingermann T: Structure of DRE, a retrotransposable element which integrates with position specificity upstream of Dictyostelium discoideum tRNA genes. Mol Cell Biol 12: 229–239 (1992).
Pearce SR, Harrison G, Li D, HeslopHarrison JS, Flavell A, Kumar A: The Ty1copia group retrotransposons in Vicia species: copy number, sequence heterogeneity and chromosomal localization. Mol Gen Genet 250: 305–315 (1996).
Saitou N, Nei M: The neighborjoining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 (1987).
SanMiguel P, Tikhonov A, Jin YK, Motchoulskaia N, Zakharov D, MelakeBerhan A, Springer PS, Edwards KJ, Lee M, Avramova Z, Bennetzen JL: Nested retrotransposons in the intergenic regions of the maize genome. Science 274: 765–768 (1996).
Schmidt T, Jung C, Metzlaff M: Distribution and evolution of two satellite DNAs in the genus Beta. Theor Appl Genet 82: 793–799 (1991)
Schmidt T, HeslopHarrison JS: Variability and evolution of highly repeated DNA sequences in the genus Beta. Genome 36: 1074–1079 (1993).
Schmidt T, Schwarzacher T, HeslopHarrison JS: Physical mapping of rRNA genes by fluorescent in situ hybridization and structural analysis of 5S rRNA genes and intergenic spacer sequences in sugar beet (Beta vulgaris). Theor Appl Genet 88: 629–636 (1994).
Schmidt T, Kubis S, HeslopHarrison JS: Analysis and chromosomal localization of retrotransposons in sugar beet (Beta vulgaris L.): LINEs and Ty1copialike elements asmajor components of the genome. Chrom Res 3: 335–345 (1995).
Schmidt T, HeslopHarrison JS: The physical and genomic organization of microsatellites in sugar beet. Proc Natl Acad Sci USA 93: 8761–8765 (1996).
SchwarzSommer Z, Leclercq L, Goebel E, Saedler H: Cin4, an insert altering the structure of the A1 gene in Zea mays, exhibits properties of nonviral retrotransposons. EMBO J 13: 3873–3880 (1987).
Voytas DF, Cummings MP, Konieczny A, Ausubel FM, Rodermel SR: copialike retrotransposons are ubiquitous among plants. Proc Natl Acad Sci USA 89: 7124–7128 (1992).
Voytas DF: Retroelements in genome organization. Science 274: 737–738 (1996).
Waugh R, McLean K, Flavell AJ, Pearce SR, Kumar A, Thomas BBT, Powell W: Genetic distribution of Bare1like retrotransposable elements in the barley genome revealed by sequencespecific amplification polymorphisms (SSAP). Mol Gen Genet 253: 687–694 (1997).
White SE, Habera LF, Wessler SR: Retrotransposons in the flanking regions of normal plant genes: A role for copialike elements in the evolution of gene structure and expression. Proc Natl Acad Sci USA 91: 11792–11796 (1994).
Wright DA, Ke N, Smalle J, Hauge BM, Goodman HM, Voytas DF: Multiple nonLTR retrotransposons in the genome of Arabidopsis thaliana. Genetics 142: 569–578 (1996).
Xiong Y, Eickbush TH: The sitespecific ribosomal DNAinsertion element R1Bm belongs to a class of nonlongterminalrepeat retrotransposons. Mol Cell Biol 8: 114–123 (1988).
Xiong Y, Eickbush TH: Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J 9: 3353–3362 (1990).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Kubis, S.E., Heslop-Harrison, J.S., Desel, C. et al. The genomic organization of non-LTR retrotransposons (LINEs) from three Beta species and five other angiosperms. Plant Mol Biol 36, 821–831 (1998). https://doi.org/10.1023/A:1005973932556
Issue Date:
DOI: https://doi.org/10.1023/A:1005973932556