Abstract
Dendritic cells (DCs) are professional antigen-presenting cells (APCs). The key functions of DCs include engulfing, processing and presenting antigens to T cells and regulating the activation of T cells. There are two major DC subtypes in human blood: plasmacytoid DCs (pDCs) and conventional DCs. To define the differences between the adult and infant immune systems, especially in terms of DC constitution, we enriched DCs from human cord blood and generated single-cell RNA sequencing data from about 7000 cells using the 10x Genomics Single Cell 3′ Solution. After incorporating the differential expression analysis method in our clustering process, we identified all the known dendritic cell subsets. Interestingly, we also found a group of DCs with gene expression that was a mix of megakaryocytes and pDCs. Further, we verified the expression of selected genes at both the RNA level by PCR and the protein level by flow cytometry. This study further demonstrates the power of single-cell RNA sequencing in dendritic cell research.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Dendritic cells (DCs) are professional antigen-presenting cells and are essential regulators of adaptive immune responses. They are derived from bone marrow and exist in both lymphoid and nonlymphoid tissues (Collin and Bigley 2018). Although DCs are small in number, as they constitute only ~1% of human peripheral blood mononuclear cells (Ginhoux et al.2018), their subsets are heterogeneous and have diverse functions. DCs are divided into three major subsets, include plasmacytoid DCs (pDCs) and two types of “conventional” or “classical” DCs (cDCs), called cDC1s and cDC2s and each subset is controlled in development by a specific repertoire of transcription factors (Merad et al.2013). pDCs produce large quantities of type I interferon upon exposure to different viruses and are also called interferon-producing cells. cDC1s are specialized for activating CD8+ T cells, while cDC2s are more potent in activating CD4+ T cells. We recently reported that a distinct CD56+ DC subpopulation exists in human blood that expresses pDC-specific surface markers (CD123 and BDCA2) but shares similar functions with cDCs (Yu et al.2015). We also found that human blood cDC2s can be further divided into two subsets (CD5high and CD5low) that differ significantly in both gene expression and function (Yin et al.2017).
Recent advances in single-cell RNA sequencing facilitate the identification of novel cell types (Jaitin et al.2014; See et al.2017; Shalek et al.2014; Villani et al.2017) and determination of cellular developmental trajectory (Zheng et al.2018). Human blood DCs have been divided into six dendritic cell types (named DC1 to DC6) by single-cell sequencing (Villani et al.2017). These six DC populations can be mapped to previously identified subtypes: DC1 corresponds to cDC1 (expresses high levels of CLEC9A and XCR1), DC2 corresponds to CD5high cDC2 (expresses high levels of CD1C)(Yin et al.2017), DC3 corresponds to CD5low cDC2 (expresses high levels of S100A8 and S100A9) (Yin et al.2017), DC5 corresponds to CD56+ DC (expresses high levels of AXL, SIGLEC6, and CD22; also called AXL+ DC) (Yu et al.2015), and DC6 corresponds to pDC (expresses high levels of CD123 and GZMB). Further, DC4 is a double-negative (CD1C−CD141−) monocyte-like cell type, which clusters with monocytes but not dendritic cells (Villani et al.2017).
The frequencies and functions of DCs are different between neonates and adults (Schuller et al.2013; Willems et al.2009; Zhang et al.2013). To further characterize neonatal DCs, we enriched dendritic cells from cord blood mononuclear cells by depleting T cells, B cells, monocytes and most NK cells and then applied single-cell RNA sequencing on the enriched cells. Comparing the resultant data with data from peripheral blood dendritic cells led us to identify all the five dendritic cell subtypes (Villani et al.2017) and a potentially novel cell type that expresses characteristic genes of both megakaryocytes and pDCs.
Results
Overview of scRNA-seq data of enriched dendritic cells from human cord blood
DCs contain heterogenous populations with diverse surface markers and functions (Yin et al.2017; Yu et al.2015; Villani et al.2017). In both adult and neonatal blood mononuclear cells, DC frequencies are low (~1%). To acquire enriched DCs from cord blood mononuclear cells (CBMCs), we labeled T cells, B cells, NK cells and monocytes with the lineages-specific markers CD3, CD19, CD16 and CD14, respectively, followed by magnetic bead depletion. Then, we applied single-cell RNA sequencing on all lineage-negative cells and performed clustering analysis and dimensional reduction.
Within the 7004 single cells which passed quality control, eight clusters (C1–C8) were identified (Fig. 1A). We mapped each cluster to known immune cell types according to a set of lineage-specific markers (Butler et al.2018) and the genes highly expressed in each cluster (Fig. 1B, and supplementary Fig. S1A, Table S1). C1, C2 and C3 cells are considered DCs that highly express HLA-DQA1 and GPR183, and these three clusters make up 61.1% of all cells. C4 cells, which express high levels of NKG5 and NKG7 (GNLY), are natural killer (NK) cells. C5, C6 and C7 cells are diverse types of progenitor cells, and they express CD34 at different levels (C5 > C6 > C7). These three clusters make up 31.3% of all the cells. Interestingly, C5 cells express FLT3, which is essential for DC development (Waskow et al.2008), suggesting that C5 may be the progenitor of DCs. Cells in C8 are erythrocytes according to their high-level expression of hemoglobin genes, such as HBA1, HBA2 and HBB (supplementary Fig. S1B).
Main clusters revealed by single-cell RNA sequencing after DC enrichment from human cord blood. A Cell clusters visualized using uniform manifold approximation and projection (UMAP). In total, eight clusters were identified and named as C1 (2804 cells), C2 (794 cells), C3 (657 cells), C4 (473 cells), C5 (1256 cells), C6 (834 cells), C7 (101 cells), and C8 (62 cells). Each dot represents an individual cell, among which the C1, C2 and C3 clusters are DCs, the C4 cluster is NK cells, the C5, C6 and C7 clusters are progenitor cells, and the C8 cluster is erythrocytes. B Heatmap showing the top five signature genes for each cluster. Clusters larger than 300 cells were randomly down sampled to 300 cells to increase the visibility of small clusters such as C7 and C8. Cells in the heatmap are ordered according to hierarchical clustering of expression profiles in each cluster. Gene significances are valued using the ROC method in the Seurat package
Currently, t-distributed stochastic neighbor embedding (t-SNE) is the most widely used method to preform dimensional reduction in single-cell analysis; this method can help to reveal local data structure in mass cytometry or single-cell transcriptomic data (Amir et al.2013). A new algorithm with similar functionality called uniform manifold approximation and projection (UMAP) was developed recently (McInnes et al.2018) and applied to biological data. UMAP has outperformed t-SNE by generating more meaningful organization of cell clusters (Becht et al.2018). To reveal whether this advantage applies to our data, we compared the UMAP output to principal component analysis (PCA, which is a well-known dimensional reduction method widely used in microarray and bulk RNA-seq analysis) and t-SNE. In the UMAP output, cell clusters identified as dendritic cells (C1, C2, and C3) grouped together in the plot, as did cell clusters identified as progenitor cells (C5, C6, and C7) (Fig. 1A). The aggregation of DC clusters and progenitor cell clusters was not observed in the PCA or t-SNE outputs (supplementary Fig. S2A, S2B). In the PCA output, different cell clusters were not well separated. In the t-SNE output, although cluster structure was clearly present, DC clusters scattered in the plot and were interwoven with progenitor cell clusters. Thus, we choose UMAP for plots for the entire analysis over t-SNE.
In droplet-based sequencing methods such as 10x, it is possible that two or more cells are captured in one droplet, thereby sharing the same cell barcode and acting like one cell during the analysis step (Zheng et al.2017). These cells are known as doublets (or multiplets), and they are minimally distinguishable from normal cells by looking at their transcriptome alone; however, they can be predicted by scanning the entire single-cell RNA-seq dataset (DePasquale et al.2018; McGinnis et al.2018; Wolock et al.2018). According to the 10x Genomics Single Cell 3′ Reagent Kits version 2 user guide, the doublet rate of about 7000 cells is approximately 5.4%. Therefore, the single-cell dataset analyzed here contains approximately 400 doublets. We predicted the identities of doublets using a newly developed method (McGinnis et al.2018) and found that the distribution of doublets did not severely skew the clustering results (supplementary Fig. S2C). However, the identities of doublets are important reference criteria during more detailed clustering analysis as follows.
Identification of known DC subtypes in scRNA-seq data
Of the three dendritic cell clusters (C1, C2, and C3), C1 and C2 express all plasmacytoid dendritic cell signature genes (e.g., GZMB and JCHAIN), while C3 shows apparent heterogeneity in DC signature gene expression (e.g., CLEC9A for cDC1 and CD1C for cDC2). It would be interesting to determine how the choice of clustering parameter influences the number of clusters in C3. The clustering parameter used in our clustering analysis pipeline (Louvain community detection method employed by the Seurat package) is called “resolution”, and it controls number of clusters indirectly; that means, as the resolution value gradually increases, new subclusters emerge and the total number of clusters increases (supplementary Fig. S3A, S3B). By calculating the differentially expressed genes between newly emerged clusters at every breaching event, we determined the optimal “resolution” value. Finally, C3 was divided into four distinct clusters (named as C3-0, C3-1, C3-2 and C3-3) (Fig. 2A, and supplementary Table S2).
DC subsets in scRNA-seq cluster C3. A Subgrouping of cluster C3. The clustering parameter (called “resolution”) was chosen according to the number of differentially expressed genes in branches of clustering tree (Fig. S2). In total, four clusters were identified and named C3-0 (306 cells), C3-1 (147 cells), C3-2 (138 cells) and C3-3 (66 cells). B Dot plot showing the expression patterns of selected genes in each subpopulation of the C3 cluster. The gene list contains the top discriminators for each dendritic cell type (Villani et al.2017). The point size corresponds to the percentage of expression cells in one group, while the color corresponds to the expression level for that gene. C Hierarchical clustering showing the relationship between subpopulations in C3. Cells in each subpopulation were pooled together, and differentially expressed genes between any two subpopulations (fold change > 1.5 and AUC > 0.7) were selected to calculate the cluster–cluster distance. D Heatmap showing expression levels of differentially expressed genes (fold change > 1.25 and AUC > 0.7) between cluster C3-0 and C3-1. E Scatter plot of normalized gene expression values (log2 TPM) for CD5high and CD5low cDC2s. Upregulated genes in C3-0 and C3-1 are highlighted in red and blue, respectively
All the dendritic cell subtypes previously identified in adult peripheral blood (Villani et al.2017; Yin et al.2017; Yu et al.2015) were found in cord blood: C3-3 is cDC1s, C3-0 and C3-1 are cDC2s, C3-2 is AXL+ DCs, and C1 and C2 are pDCs (Fig. 2B). When comparing C3-0 and C3-1, we observed that C3-1 highly expressed signature genes of CD5low cDC2s (Fig. 2B, and supplementary Fig. S2C), and the differentially expressed genes between C3-0 and C3-1 maintained their relative expression levels in bulk RNA-seq data of CD5high cDC2s and CD5low cDC2s (Yin et al.2017) (Fig. 2C, D). Although only a small portion of C3-0 cells express CD5 (7.8% in C3-0, and 3.4% in C3-1), the mean expression value of CD5 in C3-0 is higher than that in C3-1 (supplementary Fig. S3C). Therefore, we concluded that C3-0 is CD5high cDC2s, whereas C3-1 is CD5low cDC2s.
By aligning cord blood DCs and adult peripheral blood DCs together using canonical correlation analysis (CCA) (Butler et al.2018), we found that DC subtypes in cord blood matched well with DC subtypes in adult peripheral blood (supplementary Fig. S4A, S4B). In the meanwhile, DC subtypes in cord blood and adult peripheral blood shared similar signature genes (supplementary Fig. S4C, Table S3). Interestingly, AXL+ DC signature genes in cord blood are the subset of AXL+ DC signature genes in adult peripheral blood. It means some genes in AXL+ DCs (include CX3CR1 and CD5) are gradually upregulated during development. Besides, as cluster C3 cells were originally identified as cDCs, AXL+ DCs in cord blood (C3-2) are much closer to cDCs than pDCs. This relationship was also revealed by hierarchical clustering, which showed that AXL+ DCs in cord blood (C3-2) are first clustered with cDC2s and then with cDC1s (Fig. 2E).
A potentially new DC subtype with the megakaryocyte gene expression profile
In contrast to peripheral blood in which pDCs form homogeneous populations (Villani et al.2017), cord blood contains two distinct clusters of pDCs: C1 and C2. They both express pDC-specific genes such as GZMB and JCAHIN (IGJ); however, C2 also expresses megakaryocyte-specific genes such as PPBP (pro-platelet basic protein) and PF4 (platelet factor 4) (Fig. 1B, 3A, and supplementary Table S4). This indicates that cells in C1 can be matched to standard pDCs, while cells in C2 have characteristics of both pDCs and megakaryocytes. By applying the clustering method based on differential expression analysis, we found that cluster C1 cannot be further divided, while cells in cluster C2 are still further separable (supplementary Fig. S5A).
Compare C1 and C2 using pDC and megakaryocyte signature genes. A Violin plot showing the expression profile of two pDC signature genes (GZMB and JCHAIN) and two megakaryocyte signature genes (PPBP and PF4) in all dendritic cell clusters. B Dot plot showing the Spearman correlation coefficients between two pDC clusters (C1 and C2) and pure pDC and megakaryocyte clusters from public data using pDC and megakaryocyte signature genes. C Dot plot showing summed UMIs (pDC signature genes and megakaryocyte signature genes) of two pDC clusters (C1 and C2). D Heatmap showing the expression profiles of the C1 and C2 clusters. Cells in C1 are down sampled to 800 cells. Cells are ordered from left to right according to the difference between correlation to megakaryocytes and correlation to pDCs
To reveal the relationship between cluster C2 and pDCs/megakaryocytes, we used a public single-cell dataset of 33,000 peripheral blood mononuclear cells (PBMCs) which contains clusters of pDCs and megakaryocytes. To build a high-quality reference, we developed a method based on differential expression and correlation analysis to iteratively remove low-quality cells. After this cleaning step, 109 megakaryocytes and 103 pDCs were retained and then pooled together as a reference for differentially expressed genes between these two groups (supplementary Table S5). By calculating the Spearman correlation coefficients between cells in C1/C2 and the megakaryocyte/pDC reference, we found significant differences in cluster C1 and C2. All cells in C1 correlate well with pDCs, while cells in C2 form a transitional population spread between pDCs and megakaryocytes (Fig. 3B, and supplementary Fig. S5B). The diversity of cluster C2 was also revealed by the expression levels of pDC/megakaryocyte signature genes (Fig. 3C, D) and by principal component analysis (supplementary Fig. S1A). Although cluster C2 can be further divided into subgroups, we observed that the differentially expressed genes at every branching event in the clustering tree overlap with megakaryocyte signature genes (supplementary Fig. S5C). This indicates that the diversity of cluster C2 is dominated by megakaryocyte signatures.
We have several conjectures about the origin of cluster C2. One hypothesis is that these cells are “generated” during the sequencing and computational analysis steps when some megakaryocytes and pDCs form doublets (Zheng et al.2017); however, the total doublet number in the dataset (~400) is much lower than the cluster size of C2 (794). Further, if most cells in C2 are doublets between pDCs and megakaryocytes, the number of true megakaryocytes should be much larger. For further verification of the existence of C2 cells, we used flow cytometry to confirm the existence of a pDC subpopulation which expresses C2 signature genes at the cell surface. CD42a (GP9) and CLEC1B are two genes that C2 expresses at higher levels than C1, and flow cytometry analysis showed that the Lin−HLA-DR+CD1C−CD123+ pDCs indeed contain a CD42a+ or CLEC1B+ subpopulation (Fig. 4A). Compared with pDCs and cDC2s, the CLEC1B+ pDCs express higher mRNA levels of PPBP and PF4, which corresponds well with the single-cell RNA-seq data (Fig. 4B). Thus, the expressions of megakaryocyte-specific genes in C2 were confirmed at both the RNA level by PCR and the protein level by fluorescence-activated cell sorting (FACS). These experiments show that cells in cluster C2 do exist before sequencing.
Dendritic cells in the NK cells cluster
Cell heterogeneity can be observed in the heatmap of cluster signature genes (Fig. 1B). For example, a subgroup of cells in cluster C3 express signature genes of cluster C1 (pDCs), and these cells include AXL+ DC and doublets between pDCs and cDCs (Fig. S2C). Cluster C4 (NK cells) is also a heterogeneous population according to the expression of cluster C1 signature genes. To reveal whether cluster C4 contains DC-like cells, we performed the clustering analysis method previously used in C1 and C3. We found that cluster C4 can be further divided into six subpopulations (Fig. 5A).
Sub-clusters identified in C4 and compared to pDCs and NK cells. A Clustering result of cluster C4. By applying the same method used in Fig. 2A, six clusters were identified and named C4-0 (195 cells), C4-1 (92 cells), C4-2 (64 cells), C4-3 (47 cells), C4-4 (47 cells) and C4-5 (28 cells). B Violin plot showing the expression profile of two pDC signature genes (GZMB and JCHAIN) and two NK cell signature genes (NKG7 and GNLY) in all C4 subclusters. C Scatter plot showing the Spearman correlation coefficients between C4 subclusters and pure pDC/NK clusters from public data using the pDC/NK signature genes (left). Distribution of correlation coefficients between the pDC cluster and NK cell cluster (middle and right)
These six subclusters (named C4-0 to C4-5) differ significantly in pDC/NK cell signature genes. Both cluster C4-1 and C4-2 express high levels of the pDC signature genes GZMB and JCHAIN, while cluster C4-2 and C4-3 express low levels of the NK signature genes NKG7 and GNLY (Fig. 5B). To this extent, cells in cluster C4-2 are closely related to pDCs, and cells in cluster C4-1 might be doublets between pDCs and NK cells, which are supported by doublet prediction (supplementary Fig. S2C). To better explain the relationship of these subpopulations to pDCs or NK cells, we selected pDCs and NK cells from the same 33,000 PBMC single-cell dataset to build a high-quality reference (107 pDCs and 1549 NK cells were used) (supplementary Table S6). Through the use of correlation analysis, we found that cells in cluster C4-2 were more closely correlated with pDCs compared to the other C4 subpopulations (Fig. 5C). However, differential expression analysis revealed that all genes with higher expression in C4-2 cells than in C1 also had lower expression than in the C4-3 group (supplementary Fig. S6A, Table S7). One of the genes that C4-2 and C4-3 cells express at higher levels than C1 is CD3D, which is a T cell signature gene; however, C4-3 cells do not express mature T cell markers such as CD4 or CD8 (supplementary Fig. S6B). NK cells and T cells are phenotypically similar and often cluster together in single-cell analysis (Butler et al.2018; Oetjen et al.2018). Therefore, whether the C4-2 cluster is a true cell type or a mixture of pDCs and cells in C4-3 still needs to be verified.
Discussion
The single-cell RNA sequencing method provides an opportunity to scrutinize dendritic cell types in human cord blood. There are two experimental methods to analyze rare cell types such as dendritic cells: the first is to increase the total cell numbers in the single-cell dataset by pooling the sequencing results of multiple samples (Oetjen et al.2018) or integrating datasets from different sources (Butler et al.2018; Haghverdi et al.2018); the second is to increase the proportion of the specified cell type by depleting other cells before single-cell sequencing (Zheng et al.2018). We choose the second strategy of depleting major cell types (T/B/NK/Mono) in the cord blood sample. As the results show, the enrichment process was successful and over 60% of the total cells were dendritic cells.
We identified all the five dendritic cell types that have been previously identified in adult blood (Villani et al.2017; Yin et al.2017; Yu et al.2015). In contrast to adult blood (Villani et al.2017), AXL+ DCs cluster more closely with cDCs than pDCs. We also found types of pDCs in our dataset: one is a bona fide pDC (C1), and the other expresses signature genes of megakaryocytes (C2). The latter cell type has not been previously reported elsewhere. It would be interesting to know whether this cell type truly exists in vivo. Gene expression analysis showed that some cells in C2 are closely related to megakaryocytes, but others possess the characteristics of both pDCs and megakaryocytes. By applying RT-PCR and flow cytometry, we suggest that a population of pDCs expressing megakaryocyte signature genes at both the RNA and protein level does exist. Of note, platelets are fragments of cytoplasm derived from megakaryocytes. Although megakaryocytes are large (50–100 μm in diameter), platelets are much smaller (2–3 μm). Because of the common origin, the RNA expression profile of platelets and megakaryocytes is very similar (Mills et al.2017). Currently, we cannot discriminate whether C2 is an independently developed novel DC subset or just pDCs that engulfed platelets or platelet-derived vesicles. We may apply ATAC-seq (Buenrostro et al.2015) to differentiate those two possibilities. If C2 indeed expresses megakaryocyte-specific genes, the coding regions of these genes in C2 but not in C1 would be detected as open chromatin.
Doublets can result in pitfalls in single-cell analysis, and they cannot be directly distinguished from novel cell types. Two subclusters in C4 (NK cells) express pDC signature genes (C4-1 and C4-2); however, they represent an intermediate state between pDCs and another cell population (C4-0 for C4-1, C4-3 for C4-2). Doublet prediction methods support that cells in C4-1 are doublets; however, the identities of these cell populations still need to be clarified.
Materials and Methods
Cord blood purification and DC enrichment
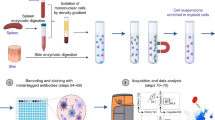
Heparinized cord blood samples processed within 24 h were collected from healthy full-term neonates (gestation time of 37 to 41 weeks) from Beijing Chaoyang Hospital Affiliated with Capital Medical University in 2018 and 2019. Cord blood mononuclear cells (CBMCs) were harvested via density gradient centrifugation using Ficoll-Paque Plus (17-1440-02, GE Healthcare). Then, CBMCs were labeled with purified mouse anti-human monoclonal antibodies against antigens including CD3, CD14, CD16 and CD19 in PBS supplemented with 2% FBS and 2 mmol/L EDTA for 30 min at 4 °C. After washing, the cells were incubated with goat anti-mouse IgG microbeads (Miltenyi Biotec) for 30 min at 4 °C. Finally, labeled cells were magnetically depleted using an LD column which was placed in the magnetic field of a midiMACS separator (Miltenyi Biotec), and the remaining cells were used for single-cell RNA sequencing.
Single-cell RNA-seq analysis
scRNA-Seq was performed using the 10x Genomics Single Cell 3′ resolution version 2 platform according to the manufacturer’s instructions. Libraries was sequenced on an Illumina X Ten apparatus and analyzed using Cell Ranger version 2.1.0 (10x Genomics). Quality control metrics were used to select cells with mitochondrial gene percentage less than 8% and with detected genes between 800 and 5500. Principle component analysis was performed, and the first 12 principle components were selected for downstream clustering and dimension reduction. Data were analyzed using the Seurat package (Version 2.3.0) in the R computational environment (Version 3.4.4). UMAP dimension reduction analysis was performed using the Python module umap-learn. Cell doublets were predicted using doubletFinder (Version 1.0.0) (McGinnis et al.2018). The number of doublets expected in the original data was set to 400.
The public 33,000 PBMCs data were downloaded as R data object from the Seurat tutorial page (https://satijalab.org/seurat/get_started_v1_4.html), which contains clustering results and cell-type assignment. Single megakaryocytes and pDCs were first selected according to the cell-type assignment in the freshly downloaded data, and then we applied a k-means-like correlation method to iteratively calculate differentially expressed genes and remove low-quality cells. In short, we first calculated the mean UMIs for pDCs and megakaryocytes and their differentially expressed genes (called gene_list_x, thresholds used were fold change > 1.5, AUC > 0.8, pct.2 < 0.2). Then, we calculated the difference between Spearman correlation coefficients to pDCs and to megakaryocytes for each cell (using gene_list_x as gene list), and cells with low difference values were removed. This process was repeated until no cells were removed. Finally, 109 megakaryocytes and 103 plasmacytoid dendritic cells were retained and used to create the cell-specific profiles. The same process was repeated to build the reference for pDCs and NK cells, in which 107 pDCs and 1549 NK cells were chosen.
Isolation of DC subsets and RT-PCR
CBMCs were washed twice with PBS containing 2% fetal bovine serum (FBS) and 2 mmol/L EDTA. Nonspecific binding was blocked using Fc receptor-blocking solution (Biolegend). The cells were stained with the following antibodies at 4 °C: anti-Lin (CD3, CD14, CD16, CD19 and CD20)-FITC, anti-CLEC1B-PE, anti-CD1c-PE-Cy7, anti-CD123-PerCP-Cy5.5, and anti-HLA-DR-APC-Cy7. Lin−HLA-DR+CD123+ CD1c− CLEC1B+ DCs were sorted using a BD FACSAria cell sorter (BD Biosciences). Detailed information about the antibodies used is presented in Table S8. cDC2 (Lin−HLA-DR+CD123−CD1c+), pDC (Lin−HLA-DR+CD123+CD1c−CLEC1B−) and CLEC1B+ DC (Lin−HLA-DR+CD123+CD1c−CLEC1B+) were sorted using a BD FACSAria cell sorter (BD Biosciences). RT-PCR was performed on the three sorted cell types, and detailed information about the primers used is stored in Table S9.
Abbreviations
- APC:
-
Antigen-presenting cell
- CB:
-
Cord blood
- CBMC:
-
Cord blood mononuclear cell
- cDC:
-
Conventional dendritic cell
- DC:
-
Dendritic cell
- FACS:
-
Fluorescence-activated cell sorting
- Mk:
-
Megakaryocyte
- Mono:
-
Monocytes
- NK cell:
-
Natural killer cell
- PBMC:
-
Peripheral blood mononuclear cell
- pDC:
-
Plasmacytoid dendritic cell
- UMI:
-
Unique molecular identifier
References
Amir ED, Davis KL, Tadmor MD, Simonds EF, Levine JH, Bendall SC, Shenfeld DK, Krishnaswamy S, Nolan GP, Pe’er D (2013) viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol 31(6):545–552
Becht E, McInnes L, Healy J, Dutertre C-A, Kwok IWH, Ng LG, Ginhoux F, Newell EW (2018) Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol 37:38–44
Buenrostro JD, Wu B, Chang HY, Greenleaf WJ (2015) ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol. https://doi.org/10.1002/0471142727.mb2129s109
Butler A, Hoffman P, Smibert P, Papalexi E, Satija R (2018) Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36(5):411–420
Collin M, Bigley V (2018) Human dendritic cell subsets: an update. Immunology 154(1):3–20
DePasquale EAK, Schnell DJ, Valiente I, Blaxall BC, Grimes HL, Singh H, Salomonis N (2018) DoubletDecon: cell-state aware removal of single-cell RNA-Seq doublets. bioRxiv 1:2. https://doi.org/10.1101/364810
Ginhoux F, See P, Lum J, Chen J (2018) A single-cell sequencing guide for immunologists. Front Immunol 9:2425
Haghverdi L, Lun ATL, Morgan MD, Marioni JC (2018) Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat Biotechnol 36(5):421–427
Jaitin DA, Kenigsberg E, Keren-Shaul H, Elefant N, Paul F, Zaretsky I, Mildner A, Cohen N, Jung S, Tanay A, Amit I (2014) Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science 343(6172):776–779
McGinnis CS, Murrow LM, Gartner ZJ (2018) DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. bioRxiv 8(4):329–337
McInnes L, Healy J, Melville J (2018) UMAP: uniform manifold approximation and projection for dimension reduction. arXiv. preprint arXiv:1802.03426
Merad M, Sathe P, Helft J, Miller J, Mortha A (2013) The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu Rev Immunol 31:563–604
Mills EW, Green R, Ingolia NT (2017) Slowed decay of mRNAs enhances platelet specific translation. Blood 129(17):e38–e48
Oetjen KA, Lindblad KE, Goswami M, Gui G, Dagur PK, Lai C, Dillon LW, McCoy JP, Hourigan CS (2018) Human bone marrow assessment by single-cell RNA sequencing, mass cytometry, and flow cytometry. JCI Insight 3(23):124928
Schuller SS, Sadeghi K, Wisgrill L, Dangl A, Diesner SC, Prusa AR, Klebermasz-Schrehof K, Greber-Platzer S, Neumuller J, Helmer H, Husslein P, Pollak A, Spittler A, Forster-Waldl E (2013) Preterm neonates display altered plasmacytoid dendritic cell function and morphology. J Leukoc Biol 93(5):781–788
See P, Dutertre C-A, Chen J, Günther P, McGovern N, Irac SE, Gunawan M, Beyer M, Händler K, Duan K, Bin Sumatoh HR, Ruffin N, Jouve M, Gea-Mallorquí E, Hennekam RCM, Lim T, Yip CC, Wen M, Malleret B, Low I, Shadan NB, Fen CFS, Tay A, Lum J, Zolezzi F, Larbi A, Poidinger M, Chan JKY, Chen Q, Rénia L, Haniffa M, Benaroch P, Schlitzer A, Schultze JL, Newell EW, Ginhoux F (2017) Mapping the human DC lineage through the integration of high-dimensional techniques. Science 356(6342):eaag3009
Shalek AK, Satija R, Shuga J, Trombetta JJ, Gennert D, Lu D, Chen P, Gertner RS, Gaublomme JT, Yosef N, Schwartz S, Fowler B, Weaver S, Wang J, Wang X, Ding R, Raychowdhury R, Friedman N, Hacohen N, Park H, May AP, Regev A (2014) Single-cell RNA-seq reveals dynamic paracrine control of cellular variation. Nature 510(7505):363–369
Villani A-C, Satija R, Reynolds G, Sarkizova S, Shekhar K, Fletcher J, Griesbeck M, Butler A, Zheng S, Lazo S, Jardine L, Dixon D, Stephenson E, Nilsson E, Grundberg I, McDonald D, Filby A, Li W, De Jager PL, Rozenblatt-Rosen O, Lane AA, Haniffa M, Regev A, Hacohen N (2017) Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 356(6335):eaag4573
Waskow C, Liu K, Darrasse-Jèze G, Guermonprez P, Ginhoux F, Merad M, Shengelia T, Yao K, Nussenzweig M (2008) The receptor tyrosine kinase Flt3 is required for dendritic cell development in peripheral lymphoid tissues. Nat Immunol 9(6):676–683
Willems F, Vollstedt S, Suter M (2009) Phenotype and function of neonatal DC. Eur J Immunol 39(1):26–35
Wolock SL, Lopez R, Klein AM (2018) Scrublet: computational identification of cell doublets in single-cell transcriptomic data. bioRxiv 8(4):281–291
Yin X, Yu H, Jin X, Li J, Guo H, Shi Q, Yin Z, Xu Y, Wang X, Liu R, Wang S, Zhang L (2017) Human blood CD1c+ dendritic cells encompass CD5high and CD5low subsets that differ significantly in phenotype, gene expression, and functions. J Immunol 198(4):1553–1564
Yu H, Zhang P, Yin X, Yin Z, Shi Q, Cui Y, Liu G, Wang S, Piccaluga PP, Jiang T, Zhang L (2015) Human BDCA2 + CD123 + CD56 + dendritic cells (DCs) related to blastic plasmacytoid dendritic cell neoplasm represent a unique myeloid DC subset. Protein Cell. 6(4):297–306
Zhang X, Lepelley A, Azria E, Lebon P, Roguet G, Schwartz O, Launay O, Leclerc C, Lo-Man R (2013) Neonatal plasmacytoid dendritic cells (pDCs) display subset variation but can elicit potent anti-viral innate responses. PLoS ONE 8(1):e52003
Zheng GXY, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, Gregory MT, Shuga J, Montesclaros L, Underwood JG, Masquelier DA, Nishimura SY, Schnall-Levin M, Wyatt PW, Hindson CM, Bharadwaj R, Wong A, Ness KD, Beppu LW, Deeg HJ, McFarland C, Loeb KR, Valente WJ, Ericson NG, Stevens EA, Radich JP, Mikkelsen TS, Hindson BJ, Bielas JH (2017) Massively parallel digital transcriptional profiling of single cells. Nat Commun 8:14049
Zheng S, Papalexi E, Butler A, Stephenson W, Satija R (2018) Molecular transitions in early progenitors during human cord blood hematopoiesis. Mol Syst Biol 14(3):e8041
Acknowledgements
This work was supported by Strategic Priority Research Program of the Chinese Academy of Sciences Grant (XDPB03) and National Natural Science Foundation of China (31670924). Xiaoyang Jin, Haisheng Yu, Shouli Wang, Guanyuan Liu and Liguo Zhang designed the project; Lingyuan Meng and Zhao Yin help to obtain cord blood samples; Xiaoyang Jin analyzed and interpreted the single-cell RNA-seq data and wrote the paper; Haisheng Yu, Linnan Zhang and Weifeng Liang performed experiments and analyzed data.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
Xiaoyang Jin, Lingyuan Meng, Zhao Yin, Haisheng Yu, Linnan Zhang, Weifeng Liang, Shouli Wang, Guanyuan Liu and Liguo Zhang declare that they have no conflicts of interest.
Ethical approval
This study was carried out with the approval of Beijing Chaoyang Hospital Affiliated with Capital Medical University Ethics Committee (2018-12-21-2).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Jin, X., Meng, L., Yin, Z. et al. Characterization of dendritic cell subtypes in human cord blood by single-cell sequencing. Biophys Rep 5, 199–208 (2019). https://doi.org/10.1007/s41048-019-00096-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s41048-019-00096-5