Abstract
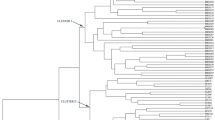
Express sequence tag (EST)-simple sequence repeat (SSR) was employed to assess the genetic diversity in elite germplasm collections of purslane (Portulaca oleracea L.) which is a saline and drought tolerant annual herb containing high amount of vitamins, minerals, and antioxidant properties. The 10 primers gave a total of ninety five bands among which 95 were polymorphic (100 %). The genetic diversity as estimated by Shannon’s information index was 1.85, revealing a high level of genetic diversity in the germplasm. The average numbers of observed allele, effective allele, expected heterozygosity, polymorphic information content (PIC), and Nei’s information index were 9.5, 5.14, 0.81, 0.33, 0.77, and 0.79, respectively. The UPGMA dendrogram based on Nei’s genetic distance grouped the whole germplasm into eight distinct clusters. The analysis of molecular variance (AMOVA) revealed that 71.87 % of total variation occurred within population while 28.13 % from among populations. Considering all these criteria and results from marker-assisted diversity analysis, accessions that are far apart based on their genetic coefficient (like Ac11 and Ac17; Ac4 and Ac42; Ac16 and Ac31; Ac38 and Ac17; Ac21 and Ac31; Ac30 and Ac10) could be selected as parents for further breeding programs.






Similar content being viewed by others
References
Alam MA, Juraimi AS, Rafii MY, Hamid AA, Aslani F, Hasan MM, Zainudin MAM, Uddin MK (2014a) Evaluation of antioxidant compounds, antioxidant activities and mineral composition of 13 collected purslane (Portulaca oleracea L.) accessions. Biomed Res Int 2014:1–10
Alam MA, Juraimi AS, Rafii MY, Hamid AA, Aslani F (2014b) Screening of Purslane (Portulaca oleracea L.) accessions for high salt tolerance. Sci World J 2014:1–12
Alam MA, Juraimi AS, Rafii MY, Hamid AA, Aslani F, Alam MZ (2014c) Effects of salinity on phenolic compounds and antioxidant activities of 13 collected purslane (Portulaca oleracea L.) germplasms. Food Chem 169:439–447
Alam MA, Juraimi AS, Rafii MY, Hamid AA, Aslani F (2014d) Collection and identification of different Purslane (Portulaca oleracea L.) accessions available in Western Peninsular Malaysia. Life Sci J 11:431–437
Anderson JA, Churchill GA, Autrique JE, Tanksley SD, Sorrells ME (1993) Optimizing parental selection for genetic linkage maps. Genome 36:181–186
Arolu IW, Rafii MY, Hanafi MM, Mahmud TMM, Latif MA (2012) Molecular characterization of Jatropha curcas germplasm using inter simple sequence repeat (ISSR) markers in Peninsular Malaysia. Aust J Crop Sci 6:1666–1673
Blair MW, González LF, Kimani PM, Butare L (2010) Genetic diversity, inter-gene pool introgression and nutritional quality of common beans (Phaseolus vulgaris L.) from Central Africa. Theor App Genet 121:237–248
Brown SM, Kresovich S (1996) The use of molecular markers in plant germplasm conservation. In: Paterson AH (ed) Genome mapping in plants, Clandes, New York, pp 85–93
Danin A, Baker I, Baker HG (1978) Cytogeography and taxonomy of the Portulaca oleracea L. polyploid complex. Israel J Bot 27:177–211
Don RH, Cox PT, Wainwright BJ, Baker K, Mattick JS (1991) ‘Touchdown’ PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res 19:4008
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Du QZ, Zhang DQ, Li BL (2012) Development of 15 novel microsatellite markers from cellulose synthase genes in Populus tomentosa (Salicaceae). Am J Bot 99:46–48
Dweck AC (2001) Purslane (Portulaca oleracea): the global panacea. Pers Care Mag 2:7–15
Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinform Online 1:47–50
Fu N, Wang P, Liu X, Shen H (2014) Use of EST-SSR markers for evaluating genetic diversity and fingerprinting Celery (Apium graveolens L.) cultivars. Molecules 19:1939–1955
Hu J, Wang L, Li J (2011) Comparison of genomic SSR and EST-SSR markers for estimating genetic diversity in cucumber. Biol Plant 55:577–580
Hwang J, Kang J, Son B, Kim K, Park Y (2011) Genetic diversity in watermelon cultivars and related species based on AFLPs and EST-SSRs. Notulae Botanicae Horti Agrobotanici Cluj-Napoca 39:285–292
Jaccard P (1908) Nouvelles recherches sur la distribution florale. Bulletin de la Société Vaudoise des Sciences Naturelles 44:223–270
Jena SN, Srivastava A, Rai KM, Ranjan A, Singh SK, Nisar T, Srivastava M, Bag SK, Mantri S, Asif MH (2012) Development and characterization of genomic and expressed SSRs for levant cotton (Gossypium herbaceum L.). Theor Appl Genet 124:565–576
Joshi SP, Gupta VS, Aggarwal RK, Ranjekar PK, Brar DS (2000) Genetic diversity and phylogenetic relationship as revealed by inter simple sequence repeat (ISSR) polymorphism in the genus Oryza. Theor Appl Genet 100:1311–1320
Kong Q, Zhang G, Chen W, Zhang Z, Zou X (2012) Identification and development of polymorphic EST-SSR markers by sequence alignment in pepper, Capsicum annuum (Solanaceae). Am J Bot 2012:59–61
Kumar H, Kaur G, Banga S (2012) Molecular characterization and assessment of genetic diversity in sesame (Sesamum indicum L.) germplasm collection using ISSR markers. J Crop Improv 26:540–557
Li G, Quiros CF (2001) Sequence-related amplified polymorphism (SRAP), a new marker system based on a simple PCR reaction: its application to mapping and gene tagging in Brassica. Theor Appl Genet 103:455–461
Li G, Ra WH, Park JW, Kwon SW, Lee JH, Park CB, Park YJ (2011) Developing EST-SSR markers to study molecular diversity in Liriope and Ophiopogon. Biochem Systemat Ecol 39:241–252
Lokhande VH, Nikam TD, Patade VY, Surasanna P (2009) Morphological and molecular diversity analysis among the Indian clones of Sesuvium portulacastrum L. Genet Res Crop Evol 56:705–717
Mujaju C, Sehic J, Nybom H (2013) Assessment of EST-SSR markers for evaluating genetic diversity in Watermelon accessions from Zimbabwe. Am J Plant Sci 4:1448–1456
Parsons JB, Newbury HT, Jackson MT, Ford-Lloyd BV (1997) Contrasting genetic diversity relationships are revealed in rice (Oryza sativa L.) using different marker types. Mole Breed 3:115–125
Ramu P, Billot C, Rami JF, Senthilvel S, Upadhyaya HD, Reddy LA, Hash CT (2013) Assessment of genetic diversity in the sorghum reference set using EST-SSR markers. Theor Appl Genet 126:2051–2064
Rao NK (2004) Plant genetic resources: advancing conservation and use through biotechnology. Afr J Biotechnol 3:136–145
Ren S, Weeda S, Akande O, Guo Y, Rutto L, Mebrahtu T (2011) Drought tolerance and AFLP-based genetic diversity in purslane (Portulaca oleracea L.). J Biotech Res 3:51–61
Rohlf FJ (2002) NTSYS-pc: numerical taxonomy system ver.2.1. Exeter Publishing Ltd., New York
Samy J, Sugumaran M, Lee KLW (2004) Herbs of Malaysia: an introduction to the medicinal, culinary, aromatic and cosmetic use of herbs. Times, Kuala Lumpur
Schulman AH (2007) Molecular markers to assess genetic diversity. Euphytica 158:313–321
Simopoulos AP, Norman HA, Gillaspy JE (1995) Purslane in human nutrition and its potential for world agriculture. World Rev Nutr Diet 77:47–74
Simopoulos AP, Tan DX, Mancheste LC, Reiter RJ (2005) Purslane: a plant source of omega-3 fatty acids and melatonin. J Pineal Res 39:331–332
Tatikonda L, Wani SP, Kannan S, Beerelli N, Sreedevi TK, Hoisington DA, Devi D, Varshney RK (2009) AFLP-based molecular characterization of an elite germplasm collection of Jatropha curcas L., a biofuel plant. Plant Sci 176:505–513
Uddin MK, Juraimi AS, Anwar F, Hossain MA, Alam MA (2012) Effect of salinity on proximate mineral composition of purslane (Portulca oleracea L.). Aust J Crop Sci 6:1732–1736
Uddin MK, Juraimi AS, Hossain MS, Nahar MAU, Ali ME, Rahman MM (2014) Purslane weed (Portulaca oleracea): A prospective plant source of nutrition, omega-3 fatty acid, and antioxidant attributes. Sci World J 2014:1–6
Varshney RK, Graner A, Sorrells ME (2005) Genetic microsatellite markers in plants: features and applications. Trends in Biotechnol 23:48–55
Vos P, Hogers R, Bleeker M, Reijans T, Van de Lee T et al (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
Wen M, Wang H, Xia Z, Zou M, Cheng LuC, Wang W (2010a) Development of EST-SSR and genomic-SSR markers to assess genetic diversity in Jatropha curcas L. BMC Res Notes 42:1–8
Wen M, Wang H, Xia Z, Zou M, Lu C, Wang W (2010b) Development of EST-SSR and genomic-SSR markers to assess genetic diversity in Jatropha Curcas L. BMC Res Notes 3:42–50
Williams JGK, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV (1990) DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res 18:6531–6535
Yan G, Aryamanesh N, Wang S (2009) Purslane – A potential vegetable crop. Rural Industries Research and Development Corporation, Australia, pp 1–26 RIRDC Publication No. 09/088
Yazici I, Turkan I, Sekmen AH, Demiral T (2007) Salinity tolerance of purslane (Portulaca oleracea L.) is achieved by enhanced antioxidative system, lower level of lipid peroxidation and proline accumulation. Environ Exp Bot 61:49–57
Yeh FC, Yang R, Boyle TJ, Ye Z, Xiyan JM (2000) PopGene32, Microsoft Windowsbased Freeware for Population Genetic Analysis, Version 1.32. Molecular Biology and Biotechnology Centre, University of Alberta, Edmonton
Zai-quan C, Fu-you Y, Ding-qing L, Teng-qiong Y, Jian F, Hui-jun Y, Qiao-fang Z, Dun-yu Z, Wei-jiao L, Xing-qi H (2012) Genetic diversity of wild rice species in Yunnan province of China. Rice Sci 19:21–28
Zhang G, Xu S, Mao W, Hu Q, Gong Y (2013) Determination of the genetic diversity of vegetable soybean [Glycine max (L.) Merr.] using EST-SSR markers. J Zhejiang Univ Sci B (Biomed & Biotechnol) 14:279–288
Zietkiewiez E, Rafalski A, Labuda D (1994) Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics 20:176–183
Acknowledgments
The authors sincerely acknowledge UPM Research University Grant (01-02-12-1695RU) for financial support of the project and IGRF (International Graduate Research Fellowship, UPM) for PhD Fellowship.
Conflict of interests
The authors declare that there is no conflict of interests regarding the publication of this article.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Alam, M.A., Juraimi, A.S., Rafii, M.Y. et al. Application of EST-SSR marker in detection of genetic variation among purslane (Portulaca oleracea L.) accessions. Braz. J. Bot 38, 119–129 (2015). https://doi.org/10.1007/s40415-014-0103-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40415-014-0103-0