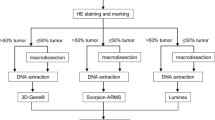
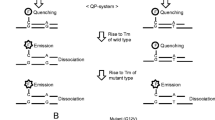
Abstract
Background and Objectives
Among patients with colorectal cancer (CRC), KRAS mutations were reported to occur in 30–51 % of all cases. CRC patients with KRAS mutations were reported to be non-responsive to anti-epidermal growth factor receptor (EGFR) monoclonal antibody (MoAb) treatment in many clinical trials. Hence, accurate detection of KRAS mutations would be critical in guiding the use of anti-EGFR MoAb therapies in CRC.
Methods
In this study, we carried out a detailed investigation of the efficacy of a wild-type (WT) blocking real-time polymerase chain reaction (PCR), employing WT KRAS locked nucleic acid blockers, and Sanger sequencing, for KRAS mutation detection in rare cells. Analyses were first conducted on cell lines to optimize the assay protocol which was subsequently applied to peripheral blood and tissue samples from patients with CRC.
Results
The optimized assay provided a superior sensitivity enabling detection of as little as two cells with mutated KRAS in the background of 104 WT cells (0.02 %). The feasibility of this assay was further investigated to assess the KRAS status of 45 colorectal tissue samples, which had been tested previously, using a conventional PCR sequencing approach. The analysis showed a mutational discordance between these two methods in 4 of 18 WT cases.
Conclusion
Our results present a simple, effective, and robust method for KRAS mutation detection in both paraffin embedded tissues and circulating tumour cells, at single-cell level. The method greatly enhances the detection sensitivity and alleviates the need of exhaustively removing co-enriched contaminating lymphocytes.




Similar content being viewed by others
References
Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127(12):2893–917.
De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11(8):753–62.
Irahara N, Baba Y, Nosho K, Shima K, Yan L, Dias-Santagata D, et al. NRAS mutations are rare in colorectal cancer. Diagn Mol Pathol. 2010;19(3):157–63.
Shen Y, Wang J, Han X, Yang H, Wang S, Lin D, et al. Effectors of epidermal growth factor receptor pathway: the genetic profiling of KRAS, BRAF, PIK3CA, NRAS mutations in colorectal cancer characteristics and personalized medicine. PLoS ONE. 2013;8(12):e81628.
Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer. 2007;7(4):295–308.
Oliveira C, Velho S, Moutinho C, Ferreira A, Preto A, Domingo E, et al. KRAS and BRAF oncogenic mutations in MSS colorectal carcinoma progression. Oncogene. 2007;26(1):158–63.
Malumbres M, Barbacid M. RAS oncogenes: the first 30 years. Nat Rev Cancer. 2003;3(6):459–65.
Shen L, Toyota M, Kondo Y, Lin E, Zhang L, Guo Y, et al. Integrated genetic and epigenetic analysis identifies three different subclasses of colon cancer. Proc Natl Acad Sci USA. 2007;104(47):18654–9.
Bos JL. Ras oncogenes in human cancer: a review. Cancer Res. 1989;49(17):4682–9.
Edkins S, O’Meara S, Parker A, Stevens C, Reis M, Jones S, et al. Recurrent KRAS codon 146 mutations in human colorectal cancer. Cancer Biol Ther. 2006;5(8):928–32.
Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3(1):11–22.
Di Fiore F, Blanchard F, Charbonnier F, Le Pessot F, Lamy A, Galais MP, et al. Clinical relevance of KRAS mutation detection in metastatic colorectal cancer treated by cetuximab plus chemotherapy. Br J Cancer. 2007;96(8):1166–9.
Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26(10):1626–34.
De Roock W, Piessevaux H, De Schutter J, Janssens M, De Hertogh G, Personeni N, et al. KRAS wild-type state predicts survival and is associated to early radiological response in metastatic colorectal cancer treated with cetuximab. Ann Oncol. 2008;19(3):508–15.
Bokemeyer C, Bondarenko I, Makhson A, Hartmann JT, Aparicio J, de Braud F, et al. Fluorouracil, leucovorin, and oxaliplatin with and without cetuximab in the first-line treatment of metastatic colorectal cancer. J Clin Oncol. 2009;27(5):663–71.
Saridaki Z, Tzardi M, Papadaki C, Sfakianaki M, Pega F, Kalikaki A, et al. Impact of KRAS, BRAF, PIK3CA mutations, PTEN, AREG, EREG expression and skin rash in ≥2 line cetuximab-based therapy of colorectal cancer patients. PLoS ONE. 2011;6(1):e15980.
Peeters M, Oliner KS, Parker A, Siena S, Van Cutsem E, Huang J, et al. Massively parallel tumor multigene sequencing to evaluate response to panitumumab in a randomized phase III study of metastatic colorectal cancer. Clin Cancer Res. 2013;19(7):1902–12.
Di Nicolantonio F, Martini M, Molinari F, Sartore-Bianchi A, Arena S, Saletti P, et al. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol. 2008;26(35):5705–12.
Garcia J, Riely GJ, Nafa K, Ladanyi M. KRAS mutational testing in the selection of patients for EGFR-targeted therapies. Semin Diagn Pathol. 2008;25(4):288–94.
Bardelli A, Siena S. Molecular mechanisms of resistance to cetuximab and panitumumab in colorectal cancer. J Clin Oncol. 2010;28(7):1254–61.
Gervasoni A, Monasterio Munoz RM, Wengler GS, Rizzi A, Zaniboni A, Parolini O. Molecular signature detection of circulating tumor cells using a panel of selected genes. Cancer Lett. 2008;263(2):267–79.
Sastre J, Maestro ML, Puente J, Veganzones S, Alfonso R, Rafael S, et al. Circulating tumor cells in colorectal cancer: correlation with clinical and pathological variables. Ann Oncol. 2008;19(5):935–8.
Zabaglo L, Ormerod MG, Parton M, Ring A, Smith IE, Dowsett M. Cell filtration-laser scanning cytometry for the characterisation of circulating breast cancer cells. Cytometry A. 2003;55(2):102–8.
Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351(8):781–91.
Zheng S, Lin H, Liu JQ, Balic M, Datar R, Cote RJ, et al. Membrane microfilter device for selective capture, electrolysis and genomic analysis of human circulating tumor cells. J Chromatogr A. 2007;1162(2):154–61.
Lim LS, Hu M, Huang MC, Cheong WC, Gan AT, Looi XL, et al. Microsieve lab-chip device for rapid enumeration and fluorescence in situ hybridization of circulating tumor cells. Lab Chip. 2012;12(21):4388–96.
Mostert B, Jiang Y, Sieuwerts AM, Wang H, Bolt-de Vries J, Biermann K, et al. KRAS and BRAF mutation status in circulating colorectal tumor cells and their correlation with primary and metastatic tumor tissue. Int J Cancer. 2013;133(1):130–41.
Sieuwerts AM, Kraan J, Bolt-de Vries J, van der Spoel P, Mostert B, Martens JW, et al. Molecular characterization of circulating tumor cells in large quantities of contaminating leukocytes by a multiplex real-time PCR. Breast Cancer Res Treat. 2009;118(3):455–68.
Newton CR, Graham A, Heptinstall LE, Powell SJ, Summers C, Kalsheker N, et al. Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS). Nucleic Acids Res. 1989;17(7):2503–16.
Hodgson DR, Clayton SJ, Girdler F, Brotherick I, Shenton B, Browell D, et al. ARMS allele-specific amplification-based detection of mutant p53 DNA and mRNA in tumors of the breast. Clin Chem. 2001;47(4):774–8.
Zuo Z, Chen SS, Chandra PK, Galbincea JM, Soape M, Doan S, et al. Application of COLD-PCR for improved detection of KRAS mutations in clinical samples. Mod Pathol. 2009;22(8):1023–31.
Pritchard CC, Akagi L, Reddy PL, Joseph L, Tait JF. COLD-PCR enhanced melting curve analysis improves diagnostic accuracy for KRAS mutations in colorectal carcinoma. BMC Clin Pathol. 2010;10:6.
Milbury CA, Correll M, Quackenbush J, Rubio R, Makrigiorgos GM. COLD-PCR enrichment of rare cancer mutations prior to targeted amplicon resequencing. Clin Chem. 2012;58(3):580–9.
Milbury CA, Li J, Makrigiorgos GM. Ice-COLD-PCR enables rapid amplification and robust enrichment for low-abundance unknown DNA mutations. Nucleic Acids Res. 2011;39(1):e2.
Castellanos-Rizaldos E, Milbury CA, Makrigiorgos GM. Enrichment of mutations in multiple DNA sequences using COLD-PCR in emulsion. PLoS ONE. 2012;7(12):e51362.
Morlan J, Baker J, Sinicropi D. Mutation detection by real-time PCR: a simple, robust and highly selective method. PLoS ONE. 2009;4(2):e4584.
Luo JD, Chan EC, Shih CL, Chen TL, Liang Y, Hwang TL, et al. Detection of rare mutant K-ras DNA in a single-tube reaction using peptide nucleic acid as both PCR clamp and sensor probe. Nucleic Acids Res. 2006;34(2):e12.
Oldenburg RP, Liu MS, Kolodney MS. Selective amplification of rare mutations using locked nucleic acid oligonucleotides that competitively inhibit primer binding to wild-type DNA. J Invest Dermatol. 2008;128(2):398–402.
Huang Q, Wang GY, Huang JF, Zhang B, Fu WL. High sensitive mutation analysis on KRAS gene using LNA/DNA chimeras as PCR amplification blockers of wild-type alleles. Mol Cell Probes. 2010;24(6):376–80.
Gasch C, Bauernhofer T, Pichler M, Langer-Freitag S, Reeh M, Seifert AM, et al. Heterogeneity of epidermal growth factor receptor status and mutations of KRAS/PIK3CA in circulating tumor cells of patients with colorectal cancer. Clin Chem. 2013;59(1):252–60.
Fabbri F, Carloni S, Zoli W, Ulivi P, Gallerani G, Fici P, et al. Detection and recovery of circulating colon cancer cells using a dielectrophoresis-based device: KRAS mutation status in pure CTCs. Cancer Lett. 2013;335(1):225–31.
Genomic DNA purification from sample applied to FTA cards. Nucleic acid sample preparation for downstream analyses principles and methods. GE Healthcare Life Sciences; 2009. p. 21–7.
Loupakis F, Ruzzo A, Cremolini C, Vincenzi B, Salvatore L, Santini D, et al. KRAS codon 61, 146 and BRAF mutations predict resistance to cetuximab plus irinotecan in KRAS codon 12 and 13 wild-type metastatic colorectal cancer. Br J Cancer. 2009;101(4):715–21.
Kopreski MS, Benko FA, Borys DJ, Khan A, McGarrity TJ, Gocke CD. Somatic mutation screening: identification of individuals harboring K-ras mutations with the use of plasma DNA. J Natl Cancer Inst. 2000;92(11):918–23.
Spindler KL, Pallisgaard N, Vogelius I, Jakobsen A. Quantitative cell-free DNA, KRAS, and BRAF mutations in plasma from patients with metastatic colorectal cancer during treatment with cetuximab and irinotecan. Clin Cancer Res. 2012;18(4):1177–85.
Acknowledgments and Disclosures
This work was funded by the Public Sector Research Funding from the Science and Engineering Research Council (grant number NR11AOT141), Agency for Science, Technology and Research (A-STAR), Singapore, and the National University Cancer Institute (NCIS) Centre Grant, Singapore (grant number NR12NMR044OM). The authors have no conflicts of interest that are directly relevant to the content of this article.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Huang, M.M.C., Leong, S.M., Chua, H.W. et al. Highly Sensitive KRAS Mutation Detection from Formalin-Fixed Paraffin-Embedded Biopsies and Circulating Tumour Cells Using Wild-Type Blocking Polymerase Chain Reaction and Sanger Sequencing. Mol Diagn Ther 18, 459–468 (2014). https://doi.org/10.1007/s40291-014-0098-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40291-014-0098-z