Abstract
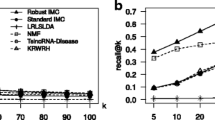
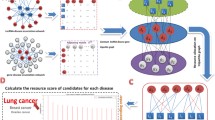
Long non-coding RNAs (lncRNAs) have been implicated in various biological processes, and are linked in many dysregulations. Over the past decade, researchers reported a large number of human disease associations with the lncRNAs, both intergenic lncRNAs (lincRNAs) and non-intergenic lncRNAs. Thanks to the next generation sequencing platform, RNA-seq, through which researchers also were able to quantify expression profiles of each of the lncRNAs in human tissue samples. In this article we adapted the non-negative matrix factorization method to develop a low-rank computational model that can describe the existing knowledge about both non-intergenic and intergenic lncRNA-disease associations represented in a two dimensional association matrix as well as convey a way of ranking disease causing lncRNAs. We proposed several NMF formulations for the problem and we found that the sparsity-constrained NMF obtained the best model among all the other models. By exploiting the inherent bi-clustering ability of the NMF models, we extracted several lncRNA groups and disease groups that possess biological significance. Moreover, we proposed an integrative NMF formulation where we incorporated along with the coding gene and lincRNA disease association data, prior knowledge about relationship networks among the coding genes and lincRNAs, and the RNA-seq expression profile data to identify potential lincRNA-coding gene co-modules with which we further enhanced the lincRNA-disease associations and untangled mysteries about functional chemistry of the intergenic lncRNAs. Experimental results show the superiority of our proposed method over two state-of-the-art clustering algorithms—k-means and hierarchical clustering.







Similar content being viewed by others
References
Bauer-Mehren A, Rautschka M, Sanz F, Furlong LI (2010) DisGeNET: a Cytoscape plugin to visualize, integrate, search and analyze gene-disease networks. Bioinformatics 26(22):2924–2926
Berry MW, Browne M, Langville AN, Pauca VP, Plemmons RJ (2007) Algorithms and applications for approximate nonnegative matrix factorization. Computat Stat Data Anal 52(1):155–173
Biswas AK, Gao JX, Zhang B, Wu X (2014) NMF-based LncRNA-disease association inference and bi-clustering. In: Proceedings of the IEEE international conference on Bioinformatics and Bioengineering (BIBE), pp 97–104
Brunet JP, Tamayo P, Golub TR, Mesirov JP (2004) Metagenes and molecular pattern discovery using matrix factorization. Proc Natl Acad Sci 101(12):4164–4169
Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL (2011) Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25(18):1915–1927
Cai D, He X, Wu X, Han J (2008) Non-negative matrix factorization on manifold. In: Data Mining. ICDM’08. Eighth IEEE International Conference on. IEEE, pp 63–72
Chen G, Wang Z, Wang D, Qiu C, Liu M, Chen X, Zhang Q, Yan G, Cui Q (2013) LncRNADisease: a database for long-non-coding RNA-associated diseases. Nucl Acids Res 41(D1):D983–D986
Chen X, Yan GY (2013) Novel human lncRNA-disease association inference based on lncRNA expression profiles. Bioinformatics 29:2617–2624
Chung S, Nakagawa H, Uemura M, Piao L, Ashikawa K, Hosono N, Takata R, Akamatsu S, Kawaguchi T, Morizono T et al (2011) Association of a novel long non-coding RNA in 8q24 with prostate cancer susceptibility. Cancer Sci 102(1):245–252
Crick F et al (1970) Central dogma of molecular biology. Nature 227(5258):561–563
Devarajan K (2008) Nonnegative matrix factorization: an analytical and interpretive tool in computational biology. PLoS Comput Biol 4(7):e1000–1029
Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci 95(25):14863–14868
Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabási AL (2007) The human disease network. Proc Natl Acad Sci 104(21):8685–8690
Gu Q, Zhou J (2009) Local learning regularized nonnegative matrix factorization. In: Twenty-First International Joint Conference on Artificial Intelligence
Guo X, Gao L, Liao Q, Xiao H, Ma X, Yang X, Luo H, Zhao G, Bu D, Jiao F et al (2013) Long non-coding RNAs function annotation: a global prediction method based on bi-colored networks. Nucl Acids Res 41(2):e35–e35
Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA (2005) Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucl Acids Res 33(suppl 1):D514–D517
Herlocker JL, Konstan JA, Terveen LG, Riedl JT (2004) Evaluating collaborative filtering recommender systems. ACM Trans Inf Syst 22(1):5–53
Hoyer PO (2004) Non-negative matrix factorization with sparseness constraints. J Mach Learn Res 5:1457–1469
Hutchins LN, Murphy SM, Singh P, Graber JH (2008) Position-dependent motif characterization using non-negative matrix factorization. Bioinformatics 24(23):2684–2690
Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermüller J, Hofacker IL et al (2007) RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316(5830):1484–1488
Kim H, Park H (2007) Sparse non-negative matrix factorizations via alternating non-negativity-constrained least squares for microarray data analysis. Bioinformatics 23(12):1495–1502
Lee DD, Seung HS (1999) Learning the parts of objects by non-negative matrix factorization. Nature 401(6755):788–791
Lee DD, Seung HS (2001) Algorithms for non-negative matrix factorization. In: Advances in neural information processing systems. pp 556–562
Li JH, Liu S, Zhou H, Qu LH, Yang JH (2013) starBase v2. 0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucl Acids Res 42:D92–D97
Liao Q, Liu C, Yuan X, Kang S, Miao R, Xiao H, Zhao G, Luo H, Bu D, Zhao H et al (2011) Large-scale prediction of long non-coding RNA functions in a coding-non-coding gene co-expression network. Nucl Acids Res 39(9):3864–3878
Lin A, Wang RT, Ahn S, Park CC, Smith DJ (2010) A genome-wide map of human genetic interactions inferred from radiation hybrid genotypes. Genome Res 20(8):1122–1132
Machado-Lima A, del Portillo HA, Durham AM (2008) Computational methods in noncoding RNA research. J Math Biol 56(1–2):15–49
Mercer TR, Dinger ME, Mattick JS (2009) Long non-coding RNAs: insights into functions. Nature Rev Genetics 10(3):155–159
Paatero P, Tapper U (1994) Positive matrix factorization: a non-negative factor model with optimal utilization of error estimates of data values. Environmetrics 5(2):111–126
Pascual-Montano A, Carazo JM, Kochi K, Lehmann D, Pascual-Marqui RD (2006) Nonsmooth nonnegative matrix factorization (nsnmf). Pattern Anal Mach Intell IEEE Trans 28(3):403–415
Pauca VP, Piper J, Plemmons RJ (2006) Nonnegative matrix factorization for spectral data analysis. Linear Algebra Appl 416(1):29–47
Prelić A, Bleuler S, Zimmermann P, Wille A, Bühlmann P, Gruissem W, Hennig L, Thiele L, Zitzler E (2006) A systematic comparison and evaluation of biclustering methods for gene expression data. Bioinformatics 22(9):1122–1129
Sacco LD, Baldassarre A, Masotti A (2011) Bioinformatics tools and novel challenges in long non-coding RNAs (lncRNAs) functional analysis. Int J Mol Sci 13(1):97–114
Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS (2010) Non-coding RNAs: regulators of disease. J Pathol 220(2):126–139
Tamayo P, Slonim D, Mesirov J, Zhu Q, Kitareewan S, Dmitrovsky E, Lander ES, Golub TR (1999) Interpreting patterns of gene expression with self-organizing maps: methods and application to hematopoietic differentiation. Proc Natl Acad Sci 96(6):2907–2912
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L (2010) Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnol 28(5):511–515
Wilusz JE, Sunwoo H, Spector DL (2009) Long noncoding RNAs: functional surprises from the RNA world. Genes Dev 23(13):1494–1504
Yang X, Gao L, Guo X, Shi X, Wu H, Song F, Wang B (2014) A network based method for analysis of lncRNA-disease associations and prediction of lncRNAs implicated in diseases. PLOS One 9(1):e87–e797
Yuan J, Wu W, Xie C, Zhao G, Zhao Y, Chen R (2014) NPInter v2. 0: an updated database of ncRNA interactions. Nucl Acids Res 42(D1):D104–D108
Zafeiriou S, Tefas A, Buciu I, Pitas I (2006) Exploiting discriminant information in nonnegative matrix factorization with application to frontal face verification. Neural Netw IEEE Trans 17(3):683–695
Zhang S, Li Q, Liu J, Zhou XJ (2011) A novel computational framework for simultaneous integration of multiple types of genomic data to identify microRNA-gene regulatory modules. Bioinformatics 27(13):i401–i409
Zhang ZY, Li T, Ding C, Ren XW, Zhang XS (2010) Binary matrix factorization for analyzing gene expression data. Data Mining Knowl Discov 20(1):28–52
Conflict of interest
The authors declare that they have no conflict of interest.
Research involving human participants and/or animals
This research neither involved any human participant nor animals. All the data-sets were collected from publicly accessible websites.
Informed consent
Not applicable to this research since no human participants, and animals were involved.
Author information
Authors and Affiliations
Corresponding author
Additional information
A short version of this paper appeared in the Proceedings of the IEEE International Conference on Bioinformatics and Bioengineering (BIBE), 2014.
Rights and permissions
About this article
Cite this article
Biswas, A.K., Kang, M., Kim, DC. et al. Inferring disease associations of the long non-coding RNAs through non-negative matrix factorization. Netw Model Anal Health Inform Bioinforma 4, 9 (2015). https://doi.org/10.1007/s13721-015-0081-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13721-015-0081-6