Abstract
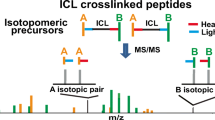
CID-MS/MS cleavable cross-linkers hold an enormous potential for an automated analysis of cross-linked products, which is essential for conducting structural proteomics studies. The created characteristic fragment ion patterns can easily be used for an automated assignment and discrimination of cross-linked products. To date, there are only a few software solutions available that make use of these properties, but none allows for an automated analysis of cleavable cross-linked products. The MeroX software fills this gap and presents a powerful tool for protein 3D-structure analysis in combination with MS/MS cleavable cross-linkers. We show that MeroX allows an automatic screening of characteristic fragment ions, considering static and variable peptide modifications, and effectively scores different types of cross-links. No manual input is required for a correct assignment of cross-links and false discovery rates are calculated. The self-explanatory graphical user interface of MeroX provides easy access for an automated cross-link search platform that is compatible with commonly used data file formats, enabling analysis of data originating from different instruments. The combination of an MS/MS cleavable cross-linker with a dedicated software tool for data analysis provides an automated workflow for 3D-structure analysis of proteins. MeroX is available at www.StavroX.com.

ᅟ







Similar content being viewed by others
References
Sinz, A.: Chemical cross-linking and mass spectrometry to map three-dimensional protein structures and protein–protein interactions. Mass Spectrom. Rev. 25, 663–682 (2006)
Young, M.M., Tang, N., Hempel, J.C., Oshiro, C.M., Taylor, E.W., Kuntz, I.D., Gibson, B.W., Dollinger, G.: High throughput protein fold identification by using experimental constraints derived from intramolecular cross-links and mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 97, 5802–5806 (2000)
Fabris, D., Yu, E.T.: Elucidating the higher-order structure of biopolymers by structural probing and mass spectrometry: MS3D. J. Mass Spectrom. 45, 841–860 (2010)
Herzog, F., Kahraman, A., Boehringer, D., Mak, R., Bracher, A., Walzthoeni, T., Leitner, A., Beck, M., Hartl, F.-U., Ban, N., Malmström, L., Aebersold, R.: Structural probing of a protein phosphatase 2A network by chemical cross-linking and mass spectrometry. Science 337, 1348–1352 (2012)
Krauth, F., Ihling, C.H., Rüttinger, H.H., Sinz, A.: Heterobifunctional isotope-labeled amine-reactive photo-cross-linker for structural investigation of proteins by matrix-assisted laser desorption/ionization tandem time-of-flight and electrospray ionization LTQ-Orbitrap mass spectrometry. Rapid Commun. Mass Spectrom. 23, 2811–2818 (2009)
Weber, P.J.A., Beck-Sickinger, A.G.: Comparison of the photochemical behavior of four different photoactivatable probes. J. Pept. Res. 49, 375–383 (1997)
Suchanek, M., Radzikowska, A., Thiele, C.: Photo-leucine and photo-methionine allow identification of protein–protein interactions in living cells. Nat. Methods 2, 261–267 (2005)
Gomes, A.F., Gozzo, F.C.: Chemical cross-linking with a diazirine photoactivatable cross-linker investigated by MALDI- and ESI-MS/MS. J. Mass Spectrom. 45, 892–899 (2010)
Kalkhof, S., Haehn, S., Paulsson, M., Smyth, N., Meiler, J., Sinz, A.: Computational modeling of laminin N-terminal domains using sparse distance constraints from disulfide bonds and chemical cross-linking. Proteins 78, 3409–3427 (2010)
Kalisman, N., Adams, C.M., Levitt, M.: Subunit order of eukaryotic TRiC/CCT chaperonin by cross-linking, mass spectrometry, and combinatorial homology modeling. Proc. Natl. Acad. Sci. U. S. A. 109, 2884–2889 (2012)
Leitner, A.: Joachimiak, Lukasz A., Bracher, A., Mönkemeyer, L., Walzthoeni, T., Chen, B., Pechmann, S., Holmes, S., Cong, Y., Ma, B., Ludtke, S., Chiu, W., Hartl, F.U., Aebersold, R., Frydman, J.: The molecular architecture of the eukaryotic chaperonin TRiC/CCT. Structure 20, 814–825 (2012)
Chen, Z.A., Jawhari, A., Fischer, L., Buchen, C., Tahir, S., Kamenski, T., Rasmussen, M., Lariviere, L., Bukowski-Wills, J.C., Nilges, M., Cramer, P., Rappsilber, J.: Architecture of the RNA polymerase II–TFIIF complex revealed by cross-linking and mass spectrometry. EMBO J. 29, 717–726 (2010)
Peri, S., Steen, H., Pandey, A.: GPMAW—a software tool for analyzing proteins and peptides. Trends Biochem. Sci. 26, 687–689 (2001)
de Koning, L.J., Kasper, P.T., Back, J.W., Nessen, M.A., Vanrobaeys, F., Van Beeumen, J., Gherardi, E., de Koster, C.G., de Jong, L.: Computer-assisted mass spectrometric analysis of naturally occurring and artificially introduced cross-links in proteins and protein complexes. FEBS J. 273, 281–291 (2006)
Rinner, O., Seebacher, J., Walzthoeni, T., Mueller, L., Beck, M., Schmidt, A., Mueller, M., Aebersold, R.: Identification of cross-linked peptides from large sequence databases. Nat. Methods 5, 315–318 (2008)
Du, X., Chowdhury, S.M., Manes, N.P., Wu, S., Mayer, M.U., Adkins, J.N., Anderson, G.A., Smith, R.D.: Xlink-Identifier: An automated data analysis platform for confident identifications of chemically cross-linked peptides using tandem mass spectrometry. J. Proteome Res. 10, 923–931 (2011)
Panchaud, A., Singh, P., Shaffer, S.A., Goodlett, D.R.: xComb: A cross-linked peptide database approach to protein − protein interaction analysis. J. Proteome Res. 9, 2508–2515 (2010)
Chu, F., Baker, P.R., Burlingame, A.L., Chalkley, R.J.: Finding chimeras: a bioinformatics strategy for identification of cross-linked peptides. Mol. Cell Proteomics 9, 25–31 (2010)
Götze, M., Pettelkau, J., Schaks, S., Bosse, K., Ihling, C., Krauth, F., Fritzsche, R., Kühn, U., Sinz, A.: StavroX—a software for analyzing crosslinked products in protein interaction studies. J. Am. Soc. Mass Spectrom. 23, 76–87 (2012)
Petrotchenko, E.V., Borchers, C.H.: ICC-CLASS: isotopically-coded cleavable crosslinking analysis software suite. BMC Bioinformatics 11, 64 (2010)
Müller, D.R., Schindler, P., Towbin, H., Wirth, U., Voshol, H., Hoving, S., Steinmetz, M.O.: Isotope-tagged cross-linking reagents. A new tool in mass spectrometric protein interaction analysis. Anal. Chem. 73, 1927–1934 (2001)
Taverner, T., Hall, N.E., O'Hair, R.A.J., Simpson, R.J.: Characterization of an antagonist interleukin-6 dimer by stable isotope labeling, cross-linking, and mass spectrometry. J. Biol. Chem. 277, 46487–46492 (2002)
Pettelkau, J., Thondorf, I., Theisgen, S., Lilie, H., Schröder, T., Arlt, C., Ihling, C.H., Sinz, A.: Structural analysis of guanylyl cyclase-activating protein-2 (GCAP-2) homodimer by stable isotope-labeling, chemical cross-linking, and mass spectrometry. J. Am. Soc. Mass Spectrom. 24, 1969–1979 (2013)
Merkley, E.D., Baker, E.S., Crowell, K.L., Orton, D.J., Taverner, T., Ansong, C., Ibrahim, Y.M., Burnet, M.C., Cort, J.R., Anderson, G.A., Smith, R.D., Adkins, J.N.: Mixed-isotope labeling with LC-IMS-MS for characterization of protein–protein interactions by chemical cross-linking. J. Am. Soc. Mass Spectrom. 24, 444–449 (2013)
Soderblom, E.J., Goshe, M.B.: Collision-induced dissociative chemical cross-linking reagents and methodology: applications to protein structural characterization using tandem mass spectrometry analysis. Anal. Chem. 78, 8059–8068 (2006)
Dreiocker, F., Müller, M.Q., Sinz, A., Schäfer, M.: Collision-induced dissociative chemical cross-linking reagent for protein structure characterization: applied Edman chemistry in the gas phase. J. Mass Spectrom. 45, 178–189 (2010)
Müller, M.Q., Dreiocker, F., Ihling, C.H., Schäfer, M., Sinz, A.: Fragmentation behavior of a thiourea-based reagent for protein structure analysis by collision-induced dissociative chemical cross-linking. J. Mass Spectrom. 45, 880–891 (2010)
Müller, M.Q., Dreiocker, F., Ihling, C.H., Schäfer, M., Sinz, A.: Cleavable cross-linker for protein structure analysis: reliable identification of cross-linking products by tandem MS. Anal. Chem. 82, 6958–6968 (2010)
Kasper, P.T., Back, J.W., Vitale, M., Hartog, A.F., Roseboom, W., de Koning, L.J., van Maarseveen, J.H., Muijsers, A.O., de Koster, C.G., de Jong, L.: An aptly positioned azido group in the spacer of a protein cross-linker for facile mapping of lysines in close proximity. Chem. Biochem. 8, 1281–1292 (2007)
Kao, A., Chiu, C.-L., Vellucci, D., Yang, Y., Patel, V.R., Guan, S., Randall, A., Baldi, P., Rychnovsky, S.D., Huang, L.: Development of a novel cross-linking strategy for fast and accurate identification of cross-linked peptides of protein complexes. Mol. Cell Proteomics 10, M110.002212 (2011)
Lu, Y., Tanasova, M., Borhan, B., Reid, G.E.: Ionic reagent for controlling the gas-phase fragmentation reactions of cross-linked peptides. Anal. Chem. 80, 9279–9287 (2008)
Lauber, M.A., Reilly, J.P.: Novel amidinating cross-linker for facilitating analyses of protein structures and interactions. Anal. Chem. 82, 7736–7743 (2010)
He, Y., Lauber, M., Reilly, J.: Unique fragmentation of singly charged DEST cross-linked peptides. J. Am. Soc. Mass Spectrom. 23, 1046–1052 (2012)
Back, J.W., Hartog, A.F., Dekker, H.L., Muijsers, A., Koning, L.J., Jong, L.: A new crosslinker for mass spectrometric analysis of the quaternary structure of protein complexes. J. Am. Soc. Mass Spectrom. 12, 222–227 (2001)
Serpa, J.J., Parker, C.E., Petrotchenko, E.V., Han, J., Pan, J., Borchers, C.H.: Mass spectrometry-based structural proteomics. Eur. J. Mass Spectrom. 18, 251–267 (2012)
Müller, M.Q., Zeiser, J.J., Dreiocker, F., Pich, A., Schäfer, M., Sinz, A.: A universal matrix-assisted laser desorption/ionization cleavable cross-linker for protein structure analysis. Rapid Commun. Mass Spectrom. 25, 155–161 (2011)
Falvo, F., Fiebig, L., Schäfer, M.: Presentation of a homobifunctional azo-reagent for protein structure analysis by collision-induced dissociative chemical cross-linking: proof-of-principle. Int. J. Mass Spectrom. 354/355, 26–32 (2013)
Schröder, T., Lilie, H., Lange, C.: The myristoylation of guanylate cyclase-activating protein-2 causes an increase in thermodynamic stability in the presence but not in the absence of Ca2+. Protein Sci. 20, 1155–1165 (2011)
Neuhoff, V., Arold, N., Taube, D., Ehrhardt, W.: Improved staining of proteins in polyacrylamide gels including isoelectric-focusing gels with clear background at nanogram sensitivity using coomassie brilliant blue G-250 and R-250. Electrophoresis 9, 255–262 (1988)
Shevchenko, A., Tomas, H., Havlis, J., Olsen, J.V., Mann, M.: In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860 (2006)
Olshevskaya, E.V., Hughes, R.E., Hurley, J.B., Dizhoor, A.M.: Calcium binding, but not a calcium-myristoyl switch, controls the ability of guanylyl cyclase-activating protein GCAP-2 to regulate photoreceptor guanylyl cyclase. J. Biol. Chem. 272, 14327–14333 (1997)
Dizhoor, A.M., Hurley, J.B.: Regulation of photoreceptor membrane guanylyl cyclases by guanylyl cyclase activator proteins. Methods 19, 521–531 (1999)
Koch, K.W., Dell'Orco, D.: A calcium-relay mechanism in vertebrate phototransduction. ACS Chem. Neurosci. 4, 909–917 (2013)
Pettelkau, J., Schröder, T., Ihling, C.H., Olausson, B.E.S., Kölbel, K., Lange, C., Sinz, A.: Structural insights into retinal guanylylcyclase-GCAP-2 interaction determined by cross-linking and mass spectrometry. Biochemistry 51, 4932–4949 (2012)
Ikura, M., Clore, G.M., Gronenborn, A.M., Zhu, G., Klee, C.B., Bax, A.: Solution structure of a calmodulin-target peptide complex by multidimensional NMR. Science 256, 632–638 (1992)
Krueger, J.K., Bishop, N.A., Blumenthal, D.K., Zhi, G., Beckingham, K., Stull, J.T., Trewhella, J.: Calmodulin binding to myosin light chain kinase begins at substoichiometric Ca2+ concentrations: a small-angle scattering study of binding and conformational transitions. Biochemistry 37, 17810–17817 (1998)
Kalkhof, S., Ihling, C., Mechtler, K., Sinz, A.: Chemical cross-linking and high-performance Fourier transform ion cyclotron resonance mass spectrometry for protein interaction analysis: application to a calmodulin/target peptide complex. Anal. Chem. 77, 495–503 (2004)
Choi, S., Jeong, J., Na, S., Lee, H.S., Kim, H.-Y., Lee, K.-J., Paek, E.: New algorithm for the identification of intact disulfide linkages based on fragmentation characteristics in tandem mass spectra. J. Proteome Res. 9, 626–635 (2009)
Frank, A.M.: Predicting intensity ranks of peptide fragment ions. J. Proteome Res. 8, 2226–2240 (2009)
Kalkhof, S., Sinz, A.: Chances and pitfalls of chemical cross-linking with amine-reactive N-hydroxysuccinimide esters. Anal. Bioanal. Chem. 392, 305–312 (2008)
Mädler, S., Bich, C., Touboul, D., Zenobi, R.: Chemical cross-linking with NHS esters: a systematic study on amino acid reactivities. J. Mass Spectrom. 44, 694–706 (2009)
Falvo, F., Fiebig, L., Dreiocker, F., Wang, R., Armentrout, P.B., Schäfer, M.: Fragmentation reactions of thiourea- and urea-compounds examined by tandem MS-, energy-resolved CID experiments, and theory. Int. J. Mass Spectrom. 330, 124–133 (2012)
Schilling, B., Row, R.H., Gibson, B.W., Guo, X., Young, M.M.: MS2Assign, automated assignment and nomenclature of tandem mass spectra of chemically crosslinked peptides. J. Am. Soc. Mass Spectrom. 14, 834–850 (2003)
Acknowledgments
This work is supported by the DFG (projects Si 867/13-1 to A.S. and SCHA 871/7-1 to M.S.) and the region of Saxony-Anhalt. M.G. is funded by the DFG (FOR855 “Cytoplasmic regulation of gene expression” and GRK1591 “Post-transcriptional control of gene expression-mechanisms and role in pathogenesis”). Francesco Falvo is acknowledged for synthesizing the urea-linker and the azo-linker, Dr. Olaf Jahn is acknowledged for synthesizing the skMLCK peptide. Dennis Glaser is acknowledged for performing the cross-linking reactions with the azo-linker. M.G. is indebted to Professor Elmar Wahle for continuous support.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Michael Götze and Jens Pettelkau contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Figure S1
SDS-PAGE analysis of GCAP-2 cross-linking reaction mixtures with the urea-linker. The cross-linking reactions were carried out in the presence (1 mM CaCl2) or in the absence (10 mM EGTA) of Ca2+. Reaction times (30, 60 min) are indicated. Marked bands were excised from the gel, digested, and analyzed by MS/MS. (DOC 5096 kb)
Supplementary Figure S2
Presentation of results by MeroX. CID fragment ion mass spectra (nano-HPLC/nano-ESI-LTQ-Orbitrap MS/MS) of (A) a doubly charged precursor ion at m/z 792.8996; (B) a triply charged precursor ion at m/z 528.9352, presenting a cross-link within GCAP-2 between Lys-178 (α-peptide, aa 177 to 182) and Lys-200 (β-peptide, aa 200-204), recorded during LC/MS analysis at 63.3-63.5 min, after reaction with the urea cross-linker (1 mM CaCl2, 30 min reaction time). Linker-specific peptide fragments modified with a butyric acid molecule are marked as Buα and Buβ, fragments modified with an isocyanate are marked as BuUrα and BuUrβ (highlighted in yellow). Backbone fragments are not labeled. (DOC 259 kb)
Supplementary Figure S3
Presentation of results by MeroX. CID fragment ion mass spectra (nano-HPLC/nano-ESI-LTQ-Orbitrap MS/MS) of (A) a doubly charged precursor ion at m/z 934.4962; (B) a triply charged precursor ion at m/z 623.3329, presenting a cross-link within GCAP-2 between Lys-129 (α-peptide, aa 129 to 136) and Lys-126 (β-peptide, aa 123-128), recorded during LC/MS analysis at 59.4-59.5 min, after reaction with the urea cross-linker (1 mM CaCl2, 30 min reaction time). Linker-specific peptide fragments modified with a butyric acid molecule are marked as Buα and Buβ, fragments modified with an isocyanate are marked as BuUrα and BuUrβ (highlighted in yellow). Backbone fragments are not labeled. (DOC 252 kb)
Supplementary Figure S4
Presentation of results by MeroX. CID fragment ion mass spectra (nano-HPLC/nano-ESI-LTQ-Orbitrap MS/MS) of (A) a doubly charged precursor ion at m/z 785.9537; (B) a triply charged precursor ion at m/z 524.3046, presenting a cross-link within GCAP-2 between Lys-126 (α-peptide, aa 123 to 128) and Lys-29 (β-peptide, aa 27-30), recorded during LC/MS analysis at 49.3-49.4 min, after reaction with the urea cross-linker (10 mM EGTA, 30 min reaction time). Linker-specific peptide fragments modified with a butyric acid molecule are marked as Buα and Buβ, fragments modified with an isocyanate are marked as BuUrα and BuUrβ (highlighted in yellow). Backbone fragments are not labeled. (DOC 258 kb)
Supplementary Figure S5
Presentation of results by MeroX. CID fragment ion mass spectra (nano-HPLC/nano-ESI-LTQ-Orbitrap MS/MS) of (A) a doubly charged precursor ion at m/z 942.5339; (B) a triply charged precursor ion at m/z 628.6912, presenting a cross-link within GCAP-2 between Lys-126 (α-peptide, aa 123 to 129) and Lys-106 (β-peptide, aa 103-108), recorded during LC/MS analysis at 40.4-40.5 min, after reaction with the urea cross-linker (1 mM CaCl2, 30 min reaction time). Linker-specific peptide fragments modified with butyric acid are marked as Buα and Buβ, fragments modified with an isocyanate are marked as BuUrα and BuUrβ (highlighted in yellow). Backbone fragments are not labeled. (DOC 254 kb)
Supplementary Figure S6
Fragment ion mass spectra of a precursor ion at m/z 954.4735 identified as CaM peptide (aa 22-37) modified with a partially hydrolyzed urea-based cross-linker molecule at Lys-31, which was detected during an LC/MS analysis at 64.8 min. (A) Manual annotation. M presents the mass of the unmodified peptide. (B) Annotation by MeroX. Fragments modified with butyric acid are marked with “+ Bu”, fragments modified with an isocyanate are marked with “+ BuUr”. (DOC 722 kb)
Supplementary Figure S7
Fragment ion mass spectra of a doubly charged precursor ion at m/z 985.4812 presenting a cross-link between CaM linked at Lys-77 (α-peptide, aa 76 to 86) and the skMLCK peptide linked at Lys-20 (β-peptide, aa 19-21), recorded in an LC/MS analysis at 31.8 min, after reaction with the urea-based cross-linker. (A) Manual annotation. Fragment ions of the α-peptide are shown in blue, fragment ions of the β-peptide are shown in red. M presents the mass of the cross-link product. (B) Annotation by MeroX. Fragments modified with γ-aminobutyric acid are marked with “+ Bu”, fragments modified with an isocyanate are marked with “+ BuUr”. The nomenclature of fragment ions is based on Schilling et al [53]. (DOC 625 kb)
Supplementary Figure S8
(A) Structure and MS/MS fragmentation of the cross-linker DSP. (B) Fragment ion mass spectra (nano-HPLC/MALDI-TOF/TOF-MS/MS) of a singly charged precursor ion at m/z 2439.1062, presenting a cross-link within GCAP-2 between Lys-50 (α-peptide, aa 48 to 62) and Lys-46 (β-peptide, aa 45-47). Linker-specific peptide fragments are marked as DT, DTSH, and DTSSH (highlighted in yellow); backbone fragments are also labeled by MeroX. (DOC 826 kb)
Supplementary Figure S9
(A) Structure and MS/MS fragmentation of the azo-linker. (B) Fragment ion mass spectra (nano-HPLC/nano-ESI-LTQ-Orbitrap MS/MS) of a triply charged precursor ion at m/z 606.6373, presenting a cross-link within GCAP-2 between Lys-50 (α-peptide, aa 48 to 56) and Lys-46 (β-peptide, aa 45-47). Linker-specific peptide fragments modified are marked as precursor (-28), corresponding to the loss of N2 (highlighted in green), Az1 and Az2 (highlighted in yellow); backbone fragments are also labeled by MeroX. (DOC 991 kb)
Supplementary Figure S10
Fragment ion mass spectra (nano-HPLC/ MALDI-TOF/TOF-MS/MS) of (A) a singly charged precursor ion at m/z 938.4428, presenting a peptide modified with a hydrolyzed urea-linker within GCAP-2 at Lys-29 (α-peptide, aa 27 to 30); “h” denotes a hydrolyzed urea-linker; (B) a singly charged precursor ion at m/z 1269.7227, presenting a cross-link within GCAP-2 between Lys-29 (α-peptide, aa 27 to 30) and Lys-46 (β-peptide, aa 45 to 47). Linker-specific peptide fragments modified with butyric acid are marked as Buα and Buβ, fragments modified with an isocyanate are marked as BuUrα and BuUrβ (highlighted in yellow). Backbone fragments are also labeled. (DOC 1077 kb)
Supplementary Table S1A
Identified GCAP-2 peptides modified by a partially hydrolyzed linker molecule (“dead-end” cross-link) (A) in the presence of 1 mM CaCl2; (B) in the presence of 10 mM EGTA. (DOC 51 kb)
Supplementary Table S1B
Identified interpeptide cross-links within GCAP-2 (A) in the presence of 1 mM CaCl2; (B) in the presence of 10 mM EGTA. (DOC 66 kb)
Supplementary Table S2A
Identified CaM peptides and skMLCK peptides modified with a partially hydrolyzed linker molecule (“dead-end” cross-link). (DOC 342 kb)
Supplementary Table S2B
Identified interpeptide cross-links within the CaM/skMLCK complex. (DOC 332 kb)
Supplementary Table S3
CaM/skMLCK peptides modified with partially hydrolyzed linker molecule (DOC 68 kb)
Supplementary Table S4
Identified cross-links within the CaM/skMLCK complex (DOC 370 kb)
Rights and permissions
About this article
Cite this article
Götze, M., Pettelkau, J., Fritzsche, R. et al. Automated Assignment of MS/MS Cleavable Cross-Links in Protein 3D-Structure Analysis. J. Am. Soc. Mass Spectrom. 26, 83–97 (2015). https://doi.org/10.1007/s13361-014-1001-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13361-014-1001-1