Abstract
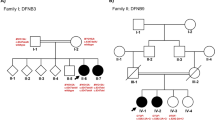
After excluding frequent mutations in common genes like GJB2, SLC26A4 and MT-RNR1 by straightforward Sanger sequencing in about 20 Polish families with hearing impairment, new and possibly pathogenic mutations were searched for by next-generation sequencing (NGS) screening using a specialised panel including more than 80 genes connected with hearing disorders. Due to high rates of false-positive pathogen predictions for newly discovered single-nucleotide polymorphisms (SNPs), different prediction models were combined to enhance the prediction power. In one family with a record of over four generations, II,3 and II,4 were suspected of hearing impairment without medical records. A male person (III,2) displayed hearing loss of 40 dB hearing level (HL) and his two sons, IV,1 and IV,2, were both affected; one with 90 dB HL and the other with 40 dB HL. Here, one heterozygous, non-synonymous variant was detected, with the SNP causing an amino acid substitution in TMC1 (transmembrane channel-like 1), a gene reported with many mutations in DFNA36 and DFNB7/11 (OMIM #606705 and #600974, respectively). Until now, the substitution p.S320R has not been described in any database. Instead of the significance of this mutation by bioinformatics tools, we confirmed the genotype–phenotype co-segregation in family members. The involvement of TMC1 in hereditary hearing impairment has not been observed in the Polish population so far.


Similar content being viewed by others
References
Adzhubei I, Jordan DM, Sunyaev SR (2013) Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet Chapter 7:Unit 7.20. doi:10.1002/0471142905.hg0720s76
Bitner-Glindzicz M (2002) Hereditary deafness and phenotyping in humans. Br Med Bull 63:73–94
Capriotti E, Fariselli P, Calabrese R, Casadio R (2005) Predicting protein stability changes from sequences using support vector machines. Bioinformatics 21(Suppl 2):ii54–ii58
De Keulenaer S, Hellemans J, Lefever S, Renard JP, De Schrijver J, Van de Voorde H, Tabatabaiefar MA, Van Nieuwerburgh F, Flamez D, Pattyn F, Scharlaken B, Deforce D, Bekaert S, Van Criekinge W, Vandesompele J, Van Camp G, Coucke P (2012) Molecular diagnostics for congenital hearing loss including 15 deafness genes using a next generation sequencing platform. BMC Med Genomics 5:17. doi:10.1186/1755-8794-5-17
Gao X, Su Y, Guan LP, Yuan YY, Huang SS, Lu Y, Wang GJ, Han MY, Yu F, Song YS, Zhu QY, Wu J, Dai P (2013) Novel compound heterozygous TMC1 mutations associated with autosomal recessive hearing loss in a Chinese family. PLoS One 8:e63026. doi:10.1371/journal.pone.0063026
Hilgert N, Alasti F, Dieltjens N, Pawlik B, Wollnik B, Uyguner O, Delmaghani S, Weil D, Petit C, Danis E, Yang T, Pandelia E, Petersen MB, Goossens D, Favero JD, Sanati MH, Smith RJ, Van Camp G (2008) Mutation analysis of TMC1 identifies four new mutations and suggests an additional deafness gene at loci DFNA36 and DFNB7/11. Clin Genet 74:223–232. doi:10.1111/j.1399-0004.2008.01053.x
Hilgert N, Monahan K, Kurima K, Li C, Friedman RA, Griffith AJ, Van Camp G (2009) Amino acid 572 in TMC1: hot spot or critical functional residue for dominant mutations causing hearing impairment. J Hum Genet 54:188–190. doi:10.1038/jhg.2009.1
Holt JR, Pan B, Koussa MA, Asai Y (2014) TMC function in hair cell transduction. Hear Res 311:17–24. doi:10.1016/j.heares.2014.01.001
Kawashima Y, Géléoc GS, Kurima K, Labay V, Lelli A, Asai Y, Makishima T, Wu DK, Della Santina CC, Holt JR, Griffith AJ (2011) Mechanotransduction in mouse inner ear hair cells requires transmembrane channel-like genes. J Clin Invest 121:4796–4809. doi:10.1172/JCI60405
Kitajiri SI, McNamara R, Makishima T, Husnain T, Zafar AU, Kittles RA, Ahmed ZM, Friedman TB, Riazuddin S, Griffith AJ (2007) Identities, frequencies and origins of TMC1 mutations causing DFNB7/B11 deafness in Pakistan. Clin Genet 72:546–550. doi:10.1111/j.1399-0004.2007.00895.x
Manji SS, Miller KA, Williams LH, Dahl HH (2012) Identification of three novel hearing loss mouse strains with mutations in the Tmc1 gene. Am J Pathol 180:1560–1569. doi:10.1016/j.ajpath.2011.12.034
Ng PC, Henikoff S (2001) Predicting deleterious amino acid substitutions. Genome Res 11:863–874. doi:10.1101/gr.176601
Nielsen M, Lundegaard C, Lund O, Petersen TN (2010) CPHmodels-3.0—remote homology modeling using structure-guided sequence profiles. Nucleic Acids Res 38:W576–W581. doi:10.1093/nar/gkq535
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi:10.1002/jcc.20084
Pollak A, Skórka A, Mueller-Malesińska M, Kostrzewa G, Kisiel B, Waligóra J, Krajewski P, Ołdak M, Korniszewski L, Skarzyński H, Ploski R (2007) M34T and V37I mutations in GJB2 associated hearing impairment: evidence for pathogenicity and reduced penetrance. Am J Med Genet A 143A:2534–2543. doi:10.1002/ajmg.a.31982
Santos RL, Wajid M, Khan MN, McArthur N, Pham TL, Bhatti A, Lee K, Irshad S, Mir A, Yan K, Chahrour MH, Ansar M, Ahmad W, Leal SM (2005) Novel sequence variants in the TMC1 gene in Pakistani families with autosomal recessive hearing impairment. Hum Mutat 26:396. doi:10.1002/humu.9374
Snoeckx RL, Huygen PL, Feldmann D, Marlin S, Denoyelle F, Waligora J, Mueller-Malesinska M, Pollak A, Ploski R, Murgia A, Orzan E, Castorina P, Ambrosetti U, Nowakowska-Szyrwinska E, Bal J, Wiszniewski W, Janecke AR, Nekahm-Heis D, Seeman P, Bendova O, Kenna MA, Frangulov A, Rehm HL, Tekin M, Incesulu A, Dahl HH, du Sart D, Jenkins L, Lucas D, Bitner-Glindzicz M, Avraham KB, Brownstein Z, del Castillo I, Moreno F, Blin N, Pfister M, Sziklai I, Toth T, Kelley PM, Cohn ES, Van Maldergem L, Hilbert P, Roux AF, Mondain M, Hoefsloot LH, Cremers CW, Löppönen T, Löppönen H, Parving A, Gronskov K, Schrijver I, Roberson J, Gualandi F, Martini A, Lina-Granade G, Pallares-Ruiz N, Correia C, Fialho G, Cryns K, Hilgert N, Van de Heyning P, Nishimura CJ, Smith RJ, Van Camp G (2005) GJB2 mutations and degree of hearing loss: a multicenter study. Am J Hum Genet 77:945–957. doi:10.1086/497996
Vreugde S, Erven A, Kros CJ, Marcotti W, Fuchs H, Kurima K, Wilcox ER, Friedman TB, Griffith AJ, Balling R, Hrabé De Angelis M, Avraham KB, Steel KP (2002) Beethoven, a mouse model for dominant, progressive hearing loss DFNA36. Nat Genet 30:257–258. doi:10.1038/ng848
Wiszniewski W, Sobieszczanska-Radoszewska L, Nowakowska-Szyrwinska E, Obersztyn E, Bal J (2001) High frequency of GJB2 gene mutations in Polish patients with prelingual nonsyndromic deafness. Genet Test 5:147–148. doi:10.1089/109065701753145637
Worth CL, Preissner R, Blundell TL (2011) SDM—a server for predicting effects of mutations on protein stability and malfunction. Nucleic Acids Res 39:W215–W222. doi:10.1093/nar/gkr363
Acknowledgments
The authors wish to acknowledge the enthusiastic cooperation of the Wroclaw hearing impairment interest group. M.A.H. was supported by a fellowship from Africa City of Technology.
Conflict of interest
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by: Michal Witt
Mohamed Ahamed Hassan and Aftab Ali Shah contributed equally to this work.
Electronic supplementary material
Below are the links to the electronic supplementary material.
Supplementary data 1
DNA sequence variants and TMC1 mutation detected in patient IV in this study using NGS, where SNPs is single nucleotide polymorphism, Ref seq is reference sequence and chrom is chromosome. (DOC 287 kb)
Supplementary data 2
Known hearing impairment genes investigated by NGS in this study. (DOC 113 kb)
Rights and permissions
About this article
Cite this article
Hassan, M.A., Shah, A.A., Szmida, E. et al. A TMC1 (transmembrane channel-like 1) mutation (p.S320R) in a Polish family with hearing impairment. J Appl Genetics 56, 311–316 (2015). https://doi.org/10.1007/s13353-014-0263-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13353-014-0263-4