Abstract
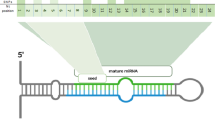
MicroRNAs (miRNAs) are a class of small, noncoding RNAs, which play a vital role in the regulation of gene expression by binding to the 3′ untranslated region (3′UTR) of a target mRNA. Despite a significant improvement in the identification of miRNAs in a variety of species, the coverage of the porcine miRNAome is still scarce. To identify porcine miRNAs potentially regulating processes taking place in the liver, we applied next generation sequencing. As a result, we detected 206 distinct miRNAs, of which 68 represented potential novel miRNAs. Among these new miRNAs, there were miRNAs deriving from the opposite arm of a hairpin precursor of already known miRNAs. Moreover, we observed 3′ and 5′ length and sequence variants, probably constituting so called isomiRs, as well as differentially mapped precursor loci, alternative precursor sequences and clustering of miRNA encoding genes. On the basis of expression levels, reflected by the number of sequence reads, we identified the most abundant miRNAs followed by gene target prediction and pathway analysis. The enriched pathways were connected with cellular and metabolic processes, growth factors as well as enzymatic activity. The obtained results are the first ones to concern the porcine liver miRNAome. Consequently, they will increase the number of known porcine miRNAs and facilitate further research on gene regulation mechanisms as well as biological processes associated with the liver functioning in pigs.

Similar content being viewed by others
References
Altuvia Y, Landgraf P, Lithwick G, Elefant N, Pfeffer S, Aravin A, Brownstein MJ, Tuschl T, Margalit H (2005) Clustering and conservation patterns of human microRNAs. Nucleic Acids Res 33:2697–2706
Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshall M, Matzke M, Ruvkun G, Tuschl T (2003) A uniform system for microRNA annotation. RNA 9:277–279
Bar M, Wyman SK, Fritz BR, Qi J, Garg KS, Parkin RK, Kroh EM, Bendoraite A, Mitchell PS, Nelson AM, Ruzzo WL, Ware C, Radich JP, Gentleman R, Ruohola-Baker H, Tewari M (2008) MicroRNA discovery and profiling in human embryonic stem cells by deep sequencing of small RNA libraries. Stem Cells 26:2496–2505
Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136:215–233
Beuvink I, Kolb FA, Budach W, Garnier A, Lange J, Natt F, Dengler U, Hall J, Filipowicz W, Weiler J (2007) A novel microarray approach reveals new tissue-specific signatures of known and predicted mammalian microRNAs. Nucleic Acids Res 35:e52
Blow MJ, Grocock RJ, van Dongen S, Enright AJ, Dicks E, Futreal PA, Wooster R, Stratton MR (2006) RNA editing of human microRNAs. Genome Biol 7:R27
Brambilla G, Cantafora A (2004) Metabolic and cardiovascular disorders in highly inbred lines for intensive pig farming: how animal welfare evaluation could improve the basic knowledge of human obesity. Ann Ist Super Sanita 40:241–244
Chen K, Rajewsky N (2006) Deep conservation of microRNAtarget relationships and 3¢UTR motifs in vertebrates, flies, and nematodes. Cold Spring Harb Symp Quant Biol 71:149–156
Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ (2005) Realtime quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 33:e179
Chin LJ, Slack FJ (2008) A truth serum for cancer-microRNAs have major potential as cancer biomarkers. Cell Res 18(10):983–984
Cho WC (2010) MicroRNAs: potential biomarkers for cancer diagnosis, prognosis and targets for therapy. Int J Biochem Cell Biol 42(8):1273–1281
Cho IS, Kim J, Seo HY, Lim do H, Hong JS, Park YH, Park DC, Hong KC, Whang KY, Lee YS (2010) Cloning and characterization of microRNAs from porcine skeletal muscle and adipose tissue. Mol Biol Rep 37(7):3567–3574
Couvelard A, Bringuier AF, Dauge MC, Nejjari M, Darai E, Benifla JL, Feldmann G, Henin D, Scoazec JY (1998) Expression of integrins during liver organogenesis in humans. Hepatology 27:839–847
Deng S, Calin GA, Croce CM, Coukos G, Zhang L (2008) Mechanisms of microRNA deregulation in human cancer. Cell Cycle 7:2643–2646
Estany J, Tor M, Villalba D, Bosch L, Gallardo D, Jiménez N, Altet L, Noguera JL, Reixach J, Amills M, Sánchez A (2007) Association of CA repeat polymorphism at intron 1 of insulin-like growth factor (IGF-I) gene with circulating IGF-I concentration, growth, and fatness in swine. Physiol Genomics 31:236–243
Fu H, Tie Y, Xu C, Zhang Z, Zhu J, Shi Y, Jiang H, Sun Z, Zheng X (2005) Identification of human fetal liver miRNAs by a novel method. FEBS Lett 579:3849–3854
Gamazon ER, Innocenti F, Wei R, Wang L, Zhang M, Mirkov S, Ramírez J, Huang RS, Cox NJ, Ratain MJ, Liu W (2013) A genome-wide integrative study of microRNAs in human liver. BMC Genomics 14:395
Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ (2006) miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 34:D140–D144
Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ (2008) miRBase: tools for microRNA genomics. Nucleic Acids Res 36:D154–D158
Hammond SM, Bernstein E, Beach D, Hannon GJ (2000) An RNAdirected nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404:293–296
He L, Hannon GJ (2004) MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 5:522–531
Jazdzewski K, Liyanarachchi S, Swierniak M, Pachucki J, Ringel MD, Jarzab B, de la Chapelle A (2009) Polymorphic mature microRNAs from passenger strand of pre-miR-146a contribute to thyroid cancer. Proc Natl Acad Sci U S A 106:1502–1505
Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ (2005) RAS is regulated by the let-7 microRNA family. Cell 120:635–647
Jones-Rhoades MW, Bartel DP, Bartel B (2006) MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol 57:19–53
Kasashima K, Nakamura Y, Kozu T (2004) Altered expression profiles of microRNAs during TPA-induced differentiation of HL-60 cells. Biochem Biophys Res Commun 322:403–410
Kim VN, Nam JW (2006) Genomics of microRNA. Trends Genet 22(3):165–173
Kim J, Cho IS, Hong JS, Choi YK, Kim H, Lee YS (2008) Identification and characterization of new microRNAs from pig. Mamm Genome 19:570–580
Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N (2005) Combinatorial microRNA target predictions. Nat Genet 37:495–500
Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T (2001) Identification of novel genes coding for small expressed RNAs. Science 294:853–858
Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foà R, Schliwka J, Fuchs U, Novosel A, Müller RU, Schermer B, Bissels U, Inman J, Phan Q, Chien M, Weir DB, Choksi R, De Vita G, Frezzetti D, Trompeter HI, Hornung V, Teng G, Hartmann G, Palkovits M, Di Lauro R, Wernet P, Macino G, Rogler CE, Nagle JW, Ju J, Papavasiliou FN, Benzing T, Lichter P, Tam W, Brownstein MJ, Bosio A, Borkhardt A, Russo JJ, Sander C, Zavolan M, Tuschl T (2007) A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129:1401–1414
Lau NC, Lim LP, Weinstein EG, Bartel DP (2001) An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294:858–862
Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN (2003) The nuclear RNase III Drosha initiates microRNA processing. Nature 425:415–419
Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN (2004) MicroRNA genes are transcribed by RNA polymerase II. EMBO J 23:4051–4060
Leung AK, Sharp PA (2007) microRNAs: a safeguard against turmoil? Cell 130(4):581–585
Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB (2003) Prediction of mammalian microRNA targets. Cell 115:787–798
Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120:15–20
Li G, Li Y, Li X, Ning X, Li M, Yang G (2011) MicroRNA identity and abundance in developing swine adipose tissue as determined by solexa sequencing. J Cell Biochem 112:1318–1328
Lu C, Tej SS, Luo S, Haudenschild CD, Meyers BC, Green PJ (2005) Elucidation of the small RNA component of the transcriptome. Science 309:1567–1569
Ma L, Teruya-Feldstein J, Weinberg RA (2007) Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 449:682–688
Meister G, Tuschl T (2004) Mechanisms of gene silencing by double-stranded RNA. Nature 431:343–349
Mi H, Lazareva-Ulitsky B, Loo R, Kejariwal A, Vandergriff J, Rabkin S, Guo N, Muruganujan A, Doremieux O, Campbell MJ, Kitano H, Thomas PD (2005) The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res 33(Database issue):D284–D2888
Morin RD, O’Connor MD, Griffith M, Kuchenbauer F, Delaney A, Prabhu AL, Zhao Y, McDonald H, Zeng T, Hirst M, Eaves CJ, Marra MA (2008) Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res 18:610–621
Moxon S, Jing R, Szittya G, Schwach F, Rusholme Pilcher RL, Moulton V, Dalmay T (2008) Deep sequencing of tomato short RNAs identifies microRNAs targeting genes involved in fruit ripening. Genome Res 18:1602–1609
Mueller DW, Bosserhoff AK (2009) Role of miRNAs in the progression of malignant melanoma. Br J Cancer 101:551–556
Nabuurs MJ (1998) Weaning piglets as a model for studying pathophysiology of diarrhea. Vet Q 20(Suppl 3):S42–S45
Natarajan A, Wagner B, Sibilia M (2007) The EGF receptor is required for efficient liver regeneration. Proc Natl Acad Sci 104:17081–17086
Neely LA, Patel S, Garver J, Gallo M, Hackett M, McLaughlin S, Nadel M, Harris J, Gullans S, Rooke J (2006) A single-molecule method for the quantitation of microRNA gene expression. Nat Methods 3:41–46
Nielsen M, Hansen JH, Hedegaard J, Nielsen RO, Panitz F, Bendixen C, Thomsen B (2010) MicroRNA identity and abundance in porcine skeletal muscles determined by deep sequencing. Anim Genet 2:159–168
Neilsen CT, Goodall GJ, Bracken CP (2012) IsomiRs—the overlooked repertoire in the dynamic microRNAome. Trends Genet 28(11):544–549
Patterson JK, Lei XG, Miller DD (2008) The pig as an experimental model for elucidating the mechanisms governing dietary influence on mineral absorption. Exp Biol Med (Maywood, NJ) 233(6):651–664
Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grasser FA, van Dyk LF, Ho CK, Shuman S, Chien M, Russo JJ, Ju J, Randall G, Lindenbach BD, Rice CM, Simon V, Ho DD, Zavolan M, Tuschl T (2005) Identification of microRNAs of the herpesvirus family. Nat Methods 2:269–276
Rajewsky N (2006) microRNA target predictions in animals. Nat Genet 38(Suppl):S8–S13
Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A (2004) Identification of mammalian microRNA host genes and transcription units. Genome Res 14:1902–1910
Rothkotter HJ (2009) Anatomical particularities of the porcine immune system-a physician’s view. Dev Comp Immunol 33(3):267–272
Sartor RB (2005) Probiotic therapy of intestinal inflammation and infections. Curr Opin Gastroenterol 21(1):44–50
Segura MF, Hanniford D, Menendez S, Reavie L, Zou X, Alvarez-Diaz S, Zakrzewski J, Blochin E, Rose A, Bogunovic D, Polsky D, Wei J, Lee P, Belitskaya-Levy I, Bhardwaj N, Osman I, Hernando E (2009) Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia associated transcription factor. Proc Natl Acad Sci U S A 106:1814–1819
Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V (2004) Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol 5(3):R13
Sharbati S, Friedländer MR, Sharbati J, Hoeke L, Chen W, Keller A, Stähler PF, Rajewsky N, Einspanier R (2010) Deciphering the porcine intestinal microRNA transcriptome. BMC Genomics 11:275
Sjogren K, Liu J-L, Blad K, Skrtic S, Vidal O, Wallenius V, LeRoith D, Tornell J, Isaksson OGP, Jansson J-O, Ohlsson C (1999) Liver-derived insulin-like growth factor I (IGF-I) is the principal source of IGF-I in blood but is not required for postnatal body growth in mice. Proc Natl Acad Sci U S A 96:7088–7092
Stocks MB, Moxon S, Mapleson D, Woolfenden HC, Mohorianu I, Folkes L, Schwach F, Dalmay T, Moulton V (2012) The UEA sRNA workbench: a suite of tools for analysing and visualizing next generation sequencing microRNA and small RNA datasets. Bioinformatics 28:2059–2061
Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, Massagué J (2008) Endogenous human microRNAs that suppress breast cancer metastasis. Nature 451:147–152
Thomson JM, Parker J, Perou CM, Hammond SM (2004) A custom microarray platform for analysis of microRNA gene expression. Nat Methods 1:47–53
Tsai WC, Hsu SD, Hsu CS, Lai TC, Chen SJ, Shen R, Huang Y, Chen HC, Lee CH, Tsai TF, Hsu MT, Wu JC, Huang HD, Shiao MS, Hsiao M, Tsou AP (2012) MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J Clin Invest 122(8):2884–2897
Tsai SM, Liu DW, Wang WP (2013) Fibroblast growth factor (Fgf) signaling pathway regulates liver homeostasis in zebrafish. Transgenic Res 22(2):301–314
Williams E, Iredale J (2000) Hepatic regeneration and TGF-beta: growing to a prosperous perfection. Gut 46(5):593–594
Vasudevan S, Tong Y, Steitz JA (2007) Switching from repression to activation: microRNAs can up-regulate translation. Science 318:1931–1934
Vaz C, Ahmad HM, Sharma P, Gupta R, Kumar L, Kulshreshtha R, Bhattacharya A (2010) Analysis of microRNA transcriptome by deep sequencing of small RNA libraries of peripheral blood. BMC Genomics 11:288
Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM (2006) A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A 103:2257–2261
Yakar S, Liu J-L, Stannard B, Butler A, Accili D, Sauer B, LeRoith D (1999) Normal growth and development in the absence of hepatic insulin-like growth factor I. Proc Natl Acad Sci U S A 96:7324–7329
Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, Calin GA, Liu CG, Croce CM, Harris CC (2006) Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 9:189–198
zuma-Mukai A, Oguri H, Mituyama T, Qian ZR, Asai K, Siomi H, Siomi MC (2008) Characterization of endogenous human Argonautes and their miRNA partners in RNA silencing. Proc Natl Acad Sci U S A 105:7964–7969
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Pawlina, K., Gurgul, A., Oczkowicz, M. et al. The characteristics of the porcine (Sus scrofa) liver miRNAome with the use of next generation sequencing. J Appl Genetics 56, 239–252 (2015). https://doi.org/10.1007/s13353-014-0245-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13353-014-0245-6