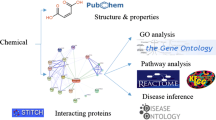
Abstract
Systems approaches are showing early promise in helping bridge the gap between pathophysiological processes and their molecular determinants. In toxicology, microarray technology leads rapid screening of DEGs (differential expressed gene) from various kinds of chemical exposes. Using toxicogenomics for the risk assessment, various and heterogeneous data are contributed to each step, such as genome sequence, genotype, gene expression, phenotype, disease information, etc. To derive actual roles of the DEGs, it is essentially required to construct interactions among DEGs and to link the known information of diseases. Proper data model is essential and critical component to build information system for risk assessment. Our study suggests a semantic modeling strategy to organize heterogeneous data types and introduces techniques and concepts (such as ontologies, semantic objects, typed relationships, contexts, graphs, and information layers) that are used to represent complex biomedical networks. We depict reconstruction of semantic relationship among chemicals, diseases, and DEGs in public available data. In this work, user’s experiment results can be easily uploaded and bound to the current data network. This feature provides to maintain user’s specific interactions from their interesting DEGs to publicly available disease and chemical data. The program was built upon DjangoWeb framework in Python language and commercial text-mining engine, MedScan, was employed. Example analysis was completed for evaluation of the system and presented in this paper. We are expecting that this work provides rapid way to build custom-driven toxico-knowledgebase by integrating customers internal documents and public data.
Similar content being viewed by others
References
Fasman, K.H., Letovsky, S.I., Cottingham, R.W. & Kingsbury, D.T. Improvements to the GDB human genome data base. Nucleic Acids Res. 24, 57–63 (1996).
Jacobson, D. & Anagnostopoulos, A. Internet resources for transgenic or targeted mutation research. Trends Genet. 12, 117–118 (1996).
Bairoch, A. The ENZYME data bank. Nucleic Acids Res. 21, 3155–3156 (1993).
Waters, M. et al. Systems toxicology and the Chemical Effects in Biological Systems (CEBS) knowledge base. EHP Toxicogenomics 111, 15–28 (2003).
Fuscoe, J.C. Impact of systems toxicology on the 3 Rs. Proc. 6th World Congress on Alternatives & Animal Use in the Life Sciences Aug 21–25, 629–632 (2007).
Shin, G.-H., Kim, H.-Y., Lee, T.-H., Park, J.-H. & Kang, B.-C. A novel semantic framework for toxicogenomics. Toxicol. Environ. Health Sci. 2, 1–3 (2010).
Sutton, P., Giudice, L.C. & Woodruff, T.J. Reproductive environmental health. Curr. Opin. Obstet. Gynecol. Dec. 22, 517–522 (2010).
Davis, A.P. et al. The Comparative Toxicogenomics Database: update 2011. Nucleic Acids Res. Jan 39 (Database issue), D1067–72 (2011).
Colborn, T., vomSaal, F.S. & Soto, A.M. Developmental effects of endocrine-disrupting chemicals in wildlife and human. Environ. Health Perspect. 101, 378–384 (1993).
vomSaal, F.S. et al. Chapel Hill bisphenol A expert panel consensus statement: integration of mechanisms, effects in animals and potential to impact human health at current levels of exposure. Reprod. Toxicol. 24. 131–138 (2007).
Saswati, P. et al. Alteration in miRNA expression profiling with response to nonylphenol in human cell lines. Mol. Cell. Toxicol. 5, 67–74 (2009).
Antizar-Ladislao, B. Environmental levels, toxicity and human exposure to tributyltin (TBT)-contaminated marine environment. Environ. Int. 34, 292–308 (2008).
Kanehisa, M. et al. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 38, D355–D360 (2010).
Degtyarenko, K. et al. ChEBI: a database and ontology for chemical entities of biological interest. Nucleic Acids Res. 36, D344–D350 (2008).
Li, Q., Wang, Y. & Bryant, S.H. A novel method for mining highly imbalanced high-throughput screening data in PubChem. Bioinformatics 25, 3310–3316 (2009).
The UniProt Consortium. Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Res. 39, D214–D219 (2011).
Python Programming Language [http://python.oyg]
Django [http://www.djangoproject.com]
SciPy [http://www.scipy.org]
Oracle DBMS [http://www.oracle.com]
MySQL DBMS [http://www.mysql.com]
SQLite DBMS [http://www.sqlite.org]
Altschul, S.F., Gish, W., Miller, W., Myers, E.W. & Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Mootha, V. et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 (2003).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kang, BC., Kim, HY., Shin, GH. et al. Semantic data integration to biological relationship among chemicals, diseases, and differential expressed genes. BioChip J 5, 63–71 (2011). https://doi.org/10.1007/s13206-011-5110-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13206-011-5110-7