Abstract
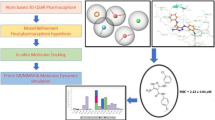
Tuberculosis is the most prominent contagious disease and needs the new targets and drugs identification. Target identification and validation is a crucial step in drug discovery process. In Mycobacterium tuberculosis, decaprenyl-phosphoryl-β-d-ribose 2′-oxidase is a potential target for antitubercular chemotherapy. It is encoded by genes dprE1 (Rv3790) and dprE2 (Rv3791). Three-dimensional (3D) structure prediction of selected target (461 amino acid residues) was intent by homology modeling using multitemplate approach based on crystal structure of 2EXR.pdb and 2Q4W.pdb with score = 48.9 bits and identities = 42/148 (29 %). A computed model was subjected to refinement based on consensus generated from two-dimensional structures and then evaluated using Structural Analysis and Verification Server to get reliable model. The final optimized model after analysis of Ramachandran plot revealed 86.5 % Core, 78.1 % Verify 3D score and 75.302 Errat values. Structure-based virtual screening against ZINC database was performed through molecular docking approach using Molegro Virtual Docker 4.2.0. The best 10 docked ligands were enumerated and validated based on their AutoDock Vina docking energy, scoring function and absorption, distribution, metabolism, excretion properties. The complex scoring function, docking energies and binding affinities revealed that these ligand molecules could be promising inhibitors against decaprenyl-phosphoryl-β-d-ribose 2′-oxidase. The present work also investigates the potential of computational molecular modeling.






Similar content being viewed by others
References
Gupta P, Hameed S, Jain R (2004) Ring-substituted imidazoles as a new class of anti-tuberculosis agents. Eur J Med Chem 39:805–814
Tangallapally RP, Sun D, Budha RN, Lee REB (2007) Discovery of novel isoxazolines as anti-tuberculosis agents. Bioorg Med Chem Lett 17:6638–6642
Tripathi R, Tewari N, Dwivedi N, Tiwari VK (2005) Fighting tuberculosis: an old disease with new challenges. Med Res Rev 25:93–131
Foroumadi A, Kiani Z, Soltani F (2003) Antituberculosis agents VIII. Synthesis and in vitro antimycobacterial activity of alkyl alpha-[5-(5-nitro-2-thienyl)-1,3,4-thiadiazole-2-ylthio] acetates. Farmaco 58:1073–1076
Medical Research Council (1948) Streptomycin treatment of pulmonary tuberculosis: Medical Research Council investigation. Br Med J 2:769–782
Pyle MM (1947) Relative numbers of resistant tubercle bacilli in sputa of patients before and during treatment with streptomycin. Proc Staff Meet Mayo Clin 22:465–473
Youmans GP, Williston EH, Feldman WH, Hinshaw HC (1946) Increase in resistance of tubercle bacilli to streptomycin: a preliminary report. Proc Staff Meet Mayo Clin 21:126–127
Pieters J (2008) Mycobacterium tuberculosis and the macrophage: maintaining a balance. Cell Host Microbe 3:399–407
Takayama K, Wang C, Besra GS (2005) Pathway to synthesis and processing of mycolic acids in Mycobacterium tuberculosis. Clin Microbiol Rev 18:81–101
Armstrong JA, Hart PD (1971) Response of cultured macrophages to Mycobacterium tuberculosis with observations on fusion of lysosomes with phagosomes. J Exp Med 134:713–740
Warner DF, Mizrahi V (2007) The survival kit of Mycobacterium tuberculosis. Nat Med 13:282–284
Neyrolles O, Hernandez PR, Pietri RF et al (2006) Is adipose tissue a place for Mycobacterium tuberculosis persistence. PLoS One 1:43
Young DB, Perkins MD, Duncan K, Barry CE (2008) Confronting the scientific obstacles to global control of tuberculosis. J Clin Invest 118:1255–1265
Goulding CW, Apostol M, Anderson DH et al (2002) The TB structural genomics consortium: providing a structural foundation for drug discovery. Curr Drug Targ Infect Disord 2:121–141
Boshoff HI, Myers TG, Copp BR et al (2002) The transcriptional responses of Mycobacterium tuberculosis to inhibitors of metabolism: novel insights into drug mechanisms of action. J Biol Chem 279:40174–40184
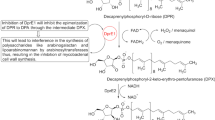
Manina G, Pasca MR, Buroni S, DeRossi E, Riccardi G (2010) Decaprenylphosphoryl-β-d-ribose 2′-epimerase from Mycobacterium tuberculosis is a magic drug target. Curr Med Chem 17:3099–3108
Mikusova K, Huang H, Yagi T et al (2005) Decaprenyl-phosphoryl arabinofuranose, the donor of the D-arabinofuranosyl residues of mycobacterial arabinan, is formed via a two-step epimerization of decaprenyl-phosphoryl ribose. J Bacteriol 187:8020–8025
Makarov V, Manina G, Mikusova K et al (2009) Benzothiazinones kill Mycobacterium tuberculosis by blocking arabinan synthesis. Science 324:801–804
Pruitt KD, Tatusova T, Klimke W, Maglott DR (2009) NCBI Reference Sequences: current status, policy and new initiatives. Nucleic Acids Res 37(Database issue):32–36
Altschul SF, Madden TL, Schäffe AAR et al (1997) Gapped BLAST and PSI BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Combet C, Blanchet C, Geourjon C, Deléage G (2000) NPS@: network protein sequence analysis. TIBS 25:147–150
Deleage G, Roux B (1987) An algorithm for protein secondary structure prediction based on class prediction. Protein Eng 4:289–294
King RD, Sternberg MJ (1996) Identification and application of the concepts important for accurate and reliable protein secondary structure prediction. Protein Sci 5:2298–2310
Gibrat JF, Garnie J, Robson B (1987) Further developments of protein secondary structure prediction using information theory: new parameters and consideration of residue pairs. J Mol Biol 5:425–443
Garnier J, Osguthorpe DJ, Robson B (1978) Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol 25:97–120
Guermeur Y (1997) Combinaison de classifieurs statistiques: application a la prediction de structure secondaire desproteines. PhD Thesis, University of Paris
Guermeur Y, Geourjon C, Gallinari P, Deleage G (1999) Improved performance in protein secondary structure prediction by inhomogeneous score combination. Bioinformatics 15:413–421
Rost B, Sander C (1993) Prediction of protein secondary structure at better than 70 % accuracy. J Mol Biol 232:584–599
Frishman D, Argos P (1996) Incorporation of non-local interactions in protein secondary structure prediction from the amino acids sequence. Protein Eng 9:133–142
Levin JM, Robson B, Garnier J (1986) An algorithm for secondary structure determination in proteins based on sequence similarity. FEBS 205:303–308
Geourjon C, Deleage G (1994) SOPM: a self-optimized method for protein secondary structure prediction. Protein Eng 7:157–164
Geourjon C, Deleage G (1995) SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput Appl Biosci 11:681–684
Rost B, Yachdav G, Liu J (2003) The predict protein server. Nuc Acids Res 32(Web Server issue):W321–W326
Zhang Y (2008) I-TASSER server for protein 3D structure prediction. BioMed Central Bioinformatics 9:40
Wu S, Zhang Y (2007) LOMETS: a local meta-threading-server for protein structure prediction. Nucleic Acids Res 35:3375–3382
Wu S, Zhang Y (2008) MUSTER: improving protein sequence profile-profile alignments by using multiple sources of structure information. Proteins struct Funct Bioinf 72:547–556
Marti RMA, Stuart A, Fiser A et al (2000) Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct 29:291–325
Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723
Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst 126:283–291
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera-a visualization system for exploratory research and analysis. J Comput Chem 25(13):1605–1612
Irwin JJ, Shoichet K (2005) ZINC-a free database of commercially available compounds for virtual screening. J Chem Inf Model 45:177–182
Khan MT, Fuskevag O, Sylte I (2009) Discovery of potent thermolysin inhibitors using structure based virtual screening and binding assay. J Med Chem 52:48–61
Castrignanò T, De Meo PD, Cozzetto D, Talamo IG, Tramontano A (2006) The PMDB protein model database. Nuc Acids Res 34(Database issue):D306–D309
Khamis MA, Gomaa W (2015) Comparative Assessment of Machine-Learning Scoring Functions on PDBbind 2013. Eng Appl Artif Intel 45:136–151
Khamis MA, Gomaa W, Fathy WA (2015) Machine Learning in Computational Docking. Artif Intell Med 63:135–152
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Anand, R. Identification of Potential Antituberculosis Drugs Through Docking and Virtual Screening. Interdiscip Sci Comput Life Sci 10, 419–429 (2018). https://doi.org/10.1007/s12539-016-0175-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12539-016-0175-6