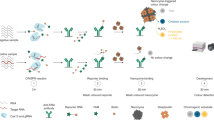
Abstract
The intrinsic affinity of DNA molecules toward metallic ions can drive the specific formation of copper nanostructures within the nucleic acid helix structure in a sequence-dependent manner. The resultant nanostructures have interesting fluorescent and electrochemical properties, which are attractive for novel biosensing applications. However, the potential of using DNA-templated nanostructures for precision disease diagnosis remains unexplored. Particularly, DNAtemplated nanostructures show high potential for the universal amplification-free detection of different RNA biomarker species. Because of their low cellular levels and differing species-dependent length and sequence features, simultaneous detection of different messenger RNAs, microRNAs, and long non-coding RNAs species with a single technique is challenging. Here, we report a contemporary technique for facile in situ assembly of DNA-templated copper nanoblocks (CuNBs) on various RNA species targets after hybridization-based magnetic isolation. Our approach circumvents the typical limitations associated with amplification and labeling procedures of current RNA assays. The synthesized CuNBs enabled amplification-free fM-level RNA detection with flexible fluorescence or electrochemical readouts. Furthermore, our nanosensing technique displays potential for clinical application, as demonstrated by non-invasive analysis of three diagnostic RNA biomarkers from a cohort of 10 prostate cancer patient urinary samples with 100%-concordance (quantitative reverse transcriptionpolymerase chain reaction (PCR) validation). The good analytical performance and versatility of our method may be useful in both diagnostics and research fields.

Similar content being viewed by others
References
Petty, J. T.; Zheng, J.; Hud, N. V.; Dickson, R. M. DNAtemplated Ag nanocluster formation. J. Am. Chem. Soc. 2004, 126, 5207–5212.
Richards, C. I.; Choi, S.; Hsiang, J. C.; Antoku, Y.; Vosch, T.; Bongiorno, A.; Tzeng, Y. L.; Dickson, R. M. Oligonucleotidestabilized Ag nanocluster fluorophores. J. Am. Chem. Soc. 2008, 130, 5038–5039.
Monson, C. F.; Woolley, A. T. DNA-templated construction of copper nanowires. Nano Lett. 2003, 3, 359–363.
Rotaru, A.; Dutta, S.; Jentzsch, E.; Gothelf, K.; Mokhir, A. Selective dsDNA-templated formation of copper nanoparticles in solution. Angew. Chem., Int. Ed. 2010, 49, 5665–5667.
Jia, X. F.; Li, J.; Han, L.; Ren, J. T.; Yang, X.; Wang, E. K. DNA-hosted copper nanoclusters for fluorescent identification of single nucleotide polymorphisms. ACS Nano 2012, 6, 3311–3317.
Qing, Z. H.; He, X. X.; He, D. G.; Wang, K. M.; Xu, F. Z.; Qing, T. P.; Yang, X. Poly(thymine)-templated selective formation of fluorescent copper nanoparticles. Angew. Chem., Int. Ed. 2013, 52, 9719–9722.
Qing, Z. H.; He, X. X.; Qing, T. P.; Wang, K. M.; Shi, H.; He, D. G.; Zou, Z.; Yan, L. A.; Xu, F. Z.; Ye, X. S.; Mao, Z. G. Poly(thymine)-templated fluorescent copper nanoparticles for ultrasensitive label-free nuclease assay and its inhibitors screening. Anal. Chem. 2013, 85, 12138–12143.
Mao, Z. G.; Qing, Z. H.; Qing, T. P.; Xu, F. Z.; Wen, L.; He, X. X.; He, D. G.; Shi, H.; Wang, K. M. Poly(thymine)- templated copper nanoparticles as a fluorescent indicator for hydrogen peroxide and oxidase-based biosensing. Anal. Chem. 2015, 87, 7454–7460.
Song, Q. W.; Shi, Y.; He, D. C.; Xu, S. H.; Ouyang, J. Sequence-dependent dsDNA-templated formation of fluorescent copper nanoparticles. Chem.—Eur. J. 2015, 21, 2417–2422.
Chen, J. Y.; Ji, X. H.; Tinnefeld, P.; He, Z. K. Multifunctional dumbbell-shaped DNA-templated selective formation of fluorescent silver nanoclusters or copper nanoparticles for sensitive detection of biomolecules. ACS Appl. Mater. Interfaces 2016, 8, 1786–1794.
Sha, L.; Zhang, X. J.; Wang, G. F. A label-free and enzyme-free ultra-sensitive transcription factors biosensor using DNA-templated copper nanoparticles as fluorescent indicator and hairpin DNA cascade reaction as signal amplifier. Biosens. Bioelectron. 2016, 82, 85–92.
Jia, X. F.; Yang, X. A.; Li, J.; Li, D. Y.; Wang, E. K. Stable Cu nanoclusters: From an aggregation-induced emission mechanism to biosensing and catalytic applications. Chem. Commun. 2014, 50, 237–239.
Brinkman, B. M. N. Splice variants as cancer biomarkers. Clin. Biochem. 2004, 37, 584–594.
Van Roosbroeck, K.; Pollet, J.; Calin, G. A. miRNAs and long noncoding RNAs as biomarkers in human diseases. Expert Rev. Mol. Diagn. 2013, 13, 183–204.
O’Leary, V. B.; Ovsepian, S. V.; Carrascosa, L. G.; Buske, F. A.; Radulovic, V.; Niyazi, M.; Moertl, S.; Trau, M.; Atkinson, M. J.; Anastasov, N. PARTICLE, a triplex-forming long ncRNA, regulates locus-specific methylation in response to low-dose irradiation. Cell Rep. 2015, 11, 474–485.
Mercer, T. R.; Dinger, M. E.; Mattick, J. S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159.
Sharp, P. A. The centrality of RNA. Cell 2009, 136, 577–580.
Riedmaier, I.; Pfaffl, M. W. Transcriptional biomarkers— High throughput screening, quantitative verification, and bioinformatical validation methods. Methods 2013, 59, 3–9.
Prensner, J. R.; Rubin, M. A.; Wei, J. T.; Chinnaiyan, A. M. Beyond PSA: The next generation of prostate cancer biomarkers. Sci. Transl. Med. 2012, 4, 127rv3.
Velonas, V. M.; Woo, H. H.; dos Remedios, C. G.; Assinder, S. J. Current status of biomarkers for prostate cancer. Int. J. Mol. Sci. 2013, 14, 11034–11060.
Fabris, L.; Ceder, Y.; Chinnaiyan, A. M.; Jenster, G. W.; Sorensen, K. D.; Tomlins, S. A.; Visakorpi, T.; Calin, G. A. The potential of microRNAs as prostate cancer biomarkers. Eur. Urol. 2016, 70, 312–322.
Rönnau, C. G. H.; Verhaegh, G. W.; Luna-Velez, M. V.; Schalken, J. A. Noncoding RNAs as novel biomarkers in prostate cancer. BioMed Res. Int. 2014, 2014, Article ID591703.
Pellegrini, K. L.; Sanda, M. G.; Moreno, C. S. RNA biomarkers to facilitate the identification of aggressive prostate cancer. Mol. Aspects Med. 2015, 45, 37–46.
Tomlins, S. A.; Bjartell, A.; Chinnaiyan, A. M.; Jenster, G.; Nam, R. K.; Rubin, M. A.; Schalken, J. A. ETS gene fusions in prostate cancer: From discovery to daily clinical practice. Eur. Urol. 2009, 56, 275–286.
Tomlins, S. A.; Rhodes, D. R.; Perner, S.; Dhanasekaran, S. M.; Mehra, R.; Sun, X. W.; Varambally, S.; Cao, X. H.; Tchinda, J.; Kuefer, R. et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 2005, 310, 644–648.
Martello, G.; Rosato, A.; Ferrari, F.; Manfrin, A.; Cordenonsi, M.; Dupont, S.; Enzo, E.; Guzzardo, V.; Rondina, M.; Spruce, T. et al. A microRNA targeting dicer for metastasis control. Cell 2010, 141, 1195–1207.
Wang, W.-X.; Kyprianou, N.; Wang, X. W.; Nelson, P. T. Dysregulation of the mitogen granulin in human cancer through the miR-15/107 microRNA gene group. Cancer Res. 2010, 70, 9137–9142.
Chen, P.-S.; Su, J.-L.; Cha, S.-T.; Tarn, W.-Y.; Wang, M.-Y.; Hsu, H.-C.; Lin, M.-T.; Chu, C.-Y.; Hua, K.-T.; Chen, C.-N. et al. miR-107 promotes tumor progression by targeting the let-7 microRNA in mice and humans. J. Clin. Investig. 2011, 121, 3442–3455.
Prensner, J. R.; Iyer, M. K.; Sahu, A.; Asangani, I. A.; Cao, Q.; Patel, L.; Vergara, I. A.; Davicioni, E.; Erho, N.; Ghadessi, M. et al. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat. Genet. 2013, 45, 1392–1398.
Hessels, D.; Smit, F. P.; Verhaegh, G. W.; Witjes, J. A.; Cornel, E. B.; Schalken, J. A. Detection of TMPRSS2-ERG fusion transcripts and prostate cancer antigen 3 in urinary sediments may improve diagnosis of prostate cancer. Clin. Cancer Res. 2007, 13, 5103–5108.
Bryant, R. J.; Pawlowski, T.; Catto, J. W. F.; Marsden, G.; Vessella, R. L.; Rhees, B.; Kuslich, C.; Visakorpi, T.; Hamdy, F. C. Changes in circulating microRNA levels associated with prostate cancer. Br. J. Cancer 2012, 106, 768–774.
Bustin, S. A. Absolute quantification of mRNA using realtime reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 2000, 25, 169–193.
Chen, C. F.; Ridzon, D. A.; Broomer, A. J.; Zhou, Z. H.; Lee, D. H.; Nguyen, J. T.; Barbisin, M.; Xu, N. L.; Mahuvakar, V. R.; Andersen, M. R. et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005, 33, e179.
Koo, K. M.; Carrascosa, L. G.; Shiddiky, M. J. A.; Trau, M. Poly(A) extensions of miRNAs for amplification-free electrochemical detection on screen-printed gold electrodes. Anal. Chem. 2016, 88, 2000–2005.
Koo, K. M.; Carrascosa, L. G.; Shiddiky, M. J. A.; Trau, M. Amplification-free detection of gene fusions in prostate cancer urinary samples using mRNA-gold affinity interactions. Anal. Chem. 2016, 88, 6781–6788.
Xu, F. Z.; Shi, H.; He, X. X.; Wang, K. M.; He, D. G.; Guo, Q. P.; Qing, Z. H.; Yan, L. A.; Ye, X. S.; Li, D. et al. Concatemeric dsDNA-templated copper nanoparticles strategy with improved sensitivity and stability based on rolling circle replication and its application in microRNA detection. Anal. Chem. 2014, 86, 6976–6982.
Wang, Z. Y.; Si, L.; Bao, J. C.; Dai, Z. H. A reusable microRNA sensor based on the electrocatalytic property of heteroduplex-templated copper nanoclusters. Chem. Commun. 2015, 51, 6305–6307.
Bakker, E.; Qin, Y. Electrochemical sensors. Anal. Chem. 2006, 78, 3965–3984.
Das, J.; Ivanov, I.; Montermini, L.; Rak, J.; Sargent, E. H.; Kelley, S. O. An electrochemical clamp assay for direct, rapid analysis of circulating nucleic acids in serum. Nat. Chem. 2015, 7, 569–575.
Gliddon, H. D.; Howes, P. D.; Kaforou, M.; Levin, M.; Stevens, M. M. A nucleic acid strand displacement system for the multiplexed detection of tuberculosis-specific mRNA using quantum dots. Nanoscale 2016, 8, 10087–10095.
Zhang, P. B.; Zhang, J. Y.; Wang, C. L.; Liu, C. H.; Wang, H.; Li, Z. P. Highly sensitive and specific multiplexed microRNA quantification using size-coded ligation chain reaction. Anal. Chem. 2014, 86, 1076–1082.
Kaffenberger, S. D.; Barbieri, C. E. Molecular subtyping of prostate cancer. Curr. Opi. Urol. 2016, 26, 213–218.
Koo, K. M.; Wee, E. J. H.; Mainwaring, P. N.; Wang, Y. L.; Trau, M. Toward precision medicine: A cancer molecular subtyping nano-strategy for RNA biomarkers in tumor and urine. Small 2016, 12, 6233–6242.
Acknowledgements
This work was supported by the UQ Postdoctoral Research Fellowship (No. 2012001456). Although not directly funding the research work in this paper, we acknowledge funding received by our laboratory from the National Breast Cancer Foundation of Australia (No. CG-12-07). This grant has significantly contributed to the environment to stimulate the research described here. K. M. K. acknowledges support from the Australian Government Research Training Program Scholarship. We thank Robert “Frank” Gardiner, Aine Farrell, Robyn Medcraft, and Clement Chow for collecting and providing clinical urine samples. We thank Junrong Li for TEM imaging.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
12274_2017_1706_MOESM1_ESM.pdf
DNA-directed assembly of copper nanoblocks with inbuilt fluorescent and electrochemical properties: Application in simultaneous amplification-free analysis of multiple RNA species
Rights and permissions
About this article
Cite this article
Koo, K.M., Carrascosa, L.G. & Trau, M. DNA-directed assembly of copper nanoblocks with inbuilt fluorescent and electrochemical properties: Application in simultaneous amplification-free analysis of multiple RNA species. Nano Res. 11, 940–952 (2018). https://doi.org/10.1007/s12274-017-1706-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12274-017-1706-0