Abstract
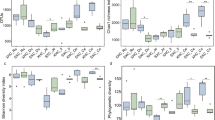
Gelatinized starch-urea (Starea, SU) is an effective and economical source of urea for ruminants. Here we assessed the influence of dietary supplementation with gelatinized starch-urea on the diversity of intestinal bacteria in finishing cattle. Fifty steers were randomly allotted to five treatments with diets supplemented with different doses of Starea [0 % (SU0), 8 % (SU8), 16 % (SU16), 24 % (SU24), and 32 % (SU32) of urea-N in total nitrogen]. Denaturing gradient gel electrophoresis (DGGE) of 16S rRNA genes was used to examine the effect of dietary supplementation of Starea on intestinal bacterial flora. Shannon–Weaver and Simpson diversity indices consistently showed the lowest bacterial diversity in the SU0 treatment. Increasing doses of Starea increased the diversity up to SU24 after which, diversity decreased. Cluster analysis of 16S rRNA gene DGGE profiles indicates that the intestinal bacterial communities associated with cattle that were not supplemented with Starea in feed differed in composition and structure from those supplemented with Starea. The amount of Starea supplemented in cattle diets influenced the abundance of several key species affiliated with Lachnospiraceae, Ruminococcaceae, Peptostreptococcaceae, Comamonadaceae and Moraxellaceae. These results suggest that Starea influences the composition and structure of intestinal bacteria which may play a role in promoting ruminant health and production performance.





Similar content being viewed by others
References
Dowd SE, Callaway TR, Wolcott RD, Sun Y, McKeehan T, Hagevoort RG, Edrington TS (2008) Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP). BMC Microbiol 8:125. doi:10.1186/1471-2180-8-125
Cummings JH, Macfarlane GT (1997) Role of intestinal bacteria in nutrient metabolism. JPEN J Parenter Enteral Nutr 21:357–365. doi:10.1177/0148607197021006357
Hooper LV, Gordon JI (2001) Commensal host–bacterial relationships in the gut. Science 292:1115–1118. doi:10.1126/science.1058709
Mapato C, Wanapat M, Cherdthong A (2010) Effects of urea treatment of straw and dietary level of vegetable oil on lactating dairy cows. Trop Anim Health Prod 42:1635–1642. doi:10.1007/s11250-010-9613-3
Taylor-Edwards CC, Hibbard G, Kitts SE, McLeod KR, Axe DE, Vanzant ES, Kristensen NB, Harmon DL (2009) Effects of slow-release urea on ruminal digesta characteristics and growth performance in beef steers. J Anim Sci 87:200–208. doi:10.2527/jas.2008-0912
Velloso L, Perry TW, Peterson RC, Beeson WM (1971) Effect of dehydrated alfalfa meal and of fish solubles on growth and nitrogen and energy balance of lambs and beef cattle fed a high urea liquid supplement. J Anim Sci 32:764–768. doi:10.2134/jas1971.324764x
Helmer LG, Bartley EE, Deyoe CW (1970) Feed processing. VI. Comparison of starea, urea, and soybean meal as protein sources for lactating dairy cows. J Dairy Sci 53:883–887. doi:10.3168/jds.S0022-0302(70)86312-3
Morrill JL, Dayton AD (1974) Soybean meal versus starea at two concentrations for young calves. J Dairy Sci 57:427–429. doi:10.3168/jds.S0022-0302(74)84908-8
Ma W, Ren L, Wang L, Ding J, Zhao J, Meng Q (2011) Effect of supplemental levels of gelatinized starch-urea on growth performance and plasma biochemical indices of growing-finishing beef cattle. Chin J Anim Nutr 23:1710–1715. doi:10.3969/j.issn.1006-267x.2011.10.010
Kim M, Kim J, Kuehn LA, Bono JL, Berry ED, Kalchayanand N, Freetly HC, Benson AK, Wells JE (2014) Investigation of bacterial diversity in the feces of cattle fed different diets. J Anim Sci 92:683–694. doi:10.2527/jas.2013-6841
Castillo-Lopez E, Ramirez Ramirez HA, Klopfenstein TJ, Anderson CL, Aluthge ND, Fernando SC, Jenkins T, Kononoff PJ (2014) Effect of feeding dried distillers grains with solubles on ruminal biohydrogenation, intestinal fatty acid profile, and gut microbial diversity evaluated through DNA pyro-sequencing. J Anim Sci 92:733–743. doi:10.2527/jas.2013-7223
Paddock ZD, Walker CE, Drouillard JS, Nagaraja TG (2011) Dietary monensin level, supplemental urea, and ractopamine on fecal shedding of Escherichia coli O157:H7 in feedlot cattle. J Anim Sci 89:2829–2835. doi:10.2527/jas.2010-3793
Rajendhran J, Gunasekaran P (2011) Microbial phylogeny and diversity: small subunit ribosomal RNA sequence analysis and beyond. Microbiol Res 166:99–110. doi:10.1016/j.micres.2010.02.003
Zhu W-Y, Williams BA, Konstantinov SR, Tamminga S, De Vos WM, Akkermans ADL (2003) Analysis of 16S rDNA reveals bacterial shift during in vitro fermentation of fermentable carbohydrate using piglet faeces as inoculum. Anaerobe 9:175–180. doi:10.1016/s1075-9964(03)00083-0
Porwal S, Lal S, Cheema S, Kalia VC (2009) Phylogeny in aid of the present and novel microbial lineages: diversity in Bacillus. PLoS ONE 4:e4438. doi:10.1371/journal.pone.0004438
Kumar P, Mehariya S, Ray S, Mishra A, Kalia VC (2014) Biodiesel industry waste: a potential source of bioenergy and biopolymers. Indian J Microbiol 55:1–7. doi:10.1007/s12088-014-0509-1
Kalia VC, Mukherjee T, Bhushan A, Joshi J, Shankar P, Huma N (2011) Analysis of the unexplored features of rrs (16S rDNA) of the Genus Clostridium. BMC Genomics 12:18. doi:10.1186/1471-2164-12-18
Lal D, Verma M, Lal R (2011) Exploring internal features of 16S rRNA gene for identification of clinically relevant species of the genus Streptococcus. Ann Clin Microbiol Antimicrob 10:28. doi:10.1186/1476-0711-10-28
Zoetendal EG, Akkermans AD, De Vos WM (1998) Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl Environ Microbiol 64:3854–3859
Nubel U, Engelen B, Felske A, Snaidr J, Wieshuber A, Amann RI, Ludwig W, Backhaus H (1996) Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J Bacteriol 178:5636–5643
Watanabe T, Asakawa S, Nakamura A, Nagaoka K, Kimura M (2004) DGGE method for analyzing 16S rDNA of methanogenic archaeal community in paddy field soil. FEMS Microbiol Lett 232:153–163. doi:10.1016/S0378-1097(04)00045-X
Wang JQ, Yin FG, Zhu C, Yu H, Niven SJ, de Lange CFM, Gong J (2012) Evaluation of probiotic bacteria for their effects on the growth performance and intestinal microbiota of newly-weaned pigs fed fermented high-moisture maize. Livestock Sci 145:79–86. doi:10.1016/j.livsci.2011.12.024
Zouache K, Raharimalala FN, Raquin V, Tran-Van V, Raveloson LH, Ravelonandro P, Mavingui P (2011) Bacterial diversity of field-caught mosquitoes, Aedes albopictus and Aedes aegypti, from different geographic regions of Madagascar. FEMS Microbiol Ecol 75:377–389. doi:10.1111/j.1574-6941.2010.01012.x
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. doi:10.1128/aem.00062-07
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi:10.1093/bioinformatics/btm404
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi:10.1093/molbev/msr121
Li S, Sun L, Wu H, Hu Z, Liu W, Li Y, Wen X (2012) The intestinal microbial diversity in mud crab (Scylla paramamosain) as determined by PCR-DGGE and clone library analysis. J Appl Microbiol 113:1341–1351. doi:10.1111/jam.12008
Haverson K, Rehakova Z, Sinkora J, Sver L, Bailey M (2007) Immune development in jejunal mucosa after colonization with selected commensal gut bacteria: a study in germ-free pigs. Vet Immunol Immunopathol 119:243–253. doi:10.1016/j.vetimm.2007.05.022
Rudi K, Moen B, Sekelja M, Frisli T, Lee MR (2012) An eight-year investigation of bovine livestock fecal microbiota. Vet Microbiol 160:369–377. doi:10.1016/j.vetmic.2012.06.003
Murphy P, Bello FD, O’Doherty JV, Arendt EK, Sweeney T, Coffey A (2012) Effects of cereal beta-glucans and enzyme inclusion on the porcine gastrointestinal tract microbiota. Anaerobe 18:557–565. doi:10.1016/j.anaerobe.2012.09.005
Dougal K, de la Fuente G, Harris PA, Girdwood SE, Pinloche E, Newbold CJ (2013) Identification of a core bacterial community within the large intestine of the horse. PLoS ONE 8:e77660. doi:10.1371/journal.pone.0077660
Scott KP, Martin JC, Duncan SH, Flint HJ (2014) Prebiotic stimulation of human colonic butyrate-producing bacteria and bifidobacteria, in vitro. FEMS Microbiol Ecol 87:30–40. doi:10.1111/1574-6941.12186
Soller JA, Schoen ME, Bartrand T, Ravenscroft JE, Ashbolt NJ (2010) Estimated human health risks from exposure to recreational waters impacted by human and non-human sources of faecal contamination. Water Res 44:4674–4691. doi:10.1016/j.watres.2010.06.049
Costa MC, Reid-Smith R, Gow S, Hannon SJ, Booker C, Rousseau J, Benedict KM, Morley PS, Weese JS (2012) Prevalence and molecular characterization of Clostridium difficile isolated from feedlot beef cattle upon arrival and mid-feeding period. BMC Vet Res 8:38. doi:10.1186/1746-6148-8-38
Boon N, Goris J, De Vos P, Verstraete W, Top EM (2001) Genetic diversity among 3-chloroaniline- and aniline-degrading strains of the Comamonadaceae. Appl Environ Microbiol 67:1107–1115. doi:10.1128/AEM.67.3.1107-1115.2001
Hamouda A, Findlay J, Al Hassan L, Amyes SG (2011) Epidemiology of Acinetobacter baumannii of animal origin. Int J Antimicrob Agents 38:314–318. doi:10.1016/j.ijantimicag.2011.06.007
Bhushan A, Joshi J, Shankar P, Kushwah J, Raju SC, Purohit HJ, Kalia VC (2013) Development of genomic tools for the identification of certain pseudomonas up to species level. Indian J Microbiol 53:253–263. doi:10.1007/s12088-013-0412-1
Acknowledgments
This study was supported by a Grant from the National Natural Science Foundation of China (No. 31172231) and the Earmarked Fund for Modern Agro-Industry Technology Research System.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Cui, Z., Meng, Q., Ma, W. et al. Diversity of the Intestinal Bacteria of Cattle Fed on Diets with Different Doses of Gelatinized Starch-Urea. Indian J Microbiol 55, 269–277 (2015). https://doi.org/10.1007/s12088-015-0526-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12088-015-0526-8