Abstract
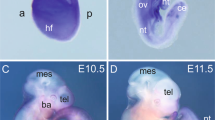
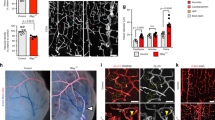
Rnf112 is a member of the RING finger protein family. The expression of Rnf112 is abundant in the brain and is regulated during brain development. Our previous study has revealed that Rnf112 can promote neuronal differentiation by inhibiting the progression of the cell cycle in cell models. In this study, we further revealed the important functions of Rnf112 in embryo development and in adult brain. Our data showed that most of the Rnf112 −/− embryos exhibited blood vascular defects and died in utero. Upon further investigation, we found that the survival rate of homozygous Rnf112 knockout mice in 129/sv and C57BL/6 mixed genetic background was increased. The survived newborns of Rnf112 −/− mice manifested growth retardation as indicated by smaller size and a reduced weight. Although the overall organization of the brain did not appear to be severely affected in Rnf112 −/− mice, using in vivo 3D MRI imaging, we found that when compared to wild-type littermates, brains of Rnf112 −/− mice were smaller. In addition, Rnf112 −/− mice displayed impairment of brain functions including motor balance, and spatial learning and memory. Our results provide important aspects for the study of Rnf112 gene functions.










Similar content being viewed by others
References
Matsuda Y, Inoue S, Seki N, Hosoi T, Orimo A et al (1996) Chromosome mapping of human (ZNF179), mouse, and rat genes for brain finger protein (bfp), a member of the RING finger family. Genomics 33:325–327
Kimura T, Arakawa Y, Inoue S, Fukushima Y, Kondo I et al (1997) The brain finger protein gene (ZNF179), a member of the RING finger family, maps within the Smith-Magenis syndrome region at 17p11.2. Am J Med Genet 69:320–324
Chen KS, Manian P, Koeuth T, Potocki L, Zhao Q et al (1997) Homologous recombination of a flanking repeat gene cluster is a mechanism for a common contiguous gene deletion syndrome. Nat Genet 17:154–163
Elsea SH, Girirajan S (2008) Smith-Magenis syndrome. Eur J Hum Genet 16:412–421
Bi W, Saifi GM, Girirajan S, Shi X, Szomju B et al (2006) RAI1 point mutations, CAG repeat variation, and SNP analysis in non-deletion Smith-Magenis syndrome. Am J Med Genet A 140:2454–2463
Zhao Q, Chen KS, Bejjani BA, Lupski JR (1998) Cloning, genomic structure, and expression of mouse ring finger protein gene Znf179. Genomics 49:394–400
Seki N, Hattori A, Muramatsu M, Saito T (1999) cDNA cloning of a human brain finger protein, BFP/ZNF179, a member of the RING finger protein family. DNA Res 6:353–356
Pao PC, Huang NK, Liu YW, Yeh SH, Lin ST et al (2011) A novel RING finger protein, Znf179, modulates cell cycle exit and neuronal differentiation of P19 embryonic stem cells. Cell Death Differ 18:1791–1804
Orimo A, Inoue S, Ikeda K, Sato M, Kato A et al (1998) Molecular cloning, localization, and developmental expression of mouse brain finger protein (Bfp)/ZNF179: distribution of bfp mRNA partially coincides with the affected areas of Smith-Magenis syndrome. Genomics 54:59–69
Deshaies RJ, Joazeiro CA (2009) RING domain E3 ubiquitin ligases. Annu Rev Biochem 78:399–434
Lomash RM, Gu X, Youle RJ, Lu W, Roche KW (2015) Neurolastin, a dynamin family GTPase, regulates excitatory synapses and spine density. Cell Rep 12:1–9
Wang SM, Lee YC, Ko CY, Lai MD, Lin DY et al (2015) Increase of zinc finger protein 179 in response to CCAAT/enhancer binding protein delta conferring an antiapoptotic effect in astrocytes of Alzheimer’s disease. Mol Neurobiol 51:370–382
Morton AJ, Hunt MJ, Hodges AK, Lewis PD, Redfern AJ et al (2005) A combination drug therapy improves cognition and reverses gene expression changes in a mouse model of Huntington’s disease. Eur J Neurosci 21:855–870
Ferraiuolo L, Heath PR, Holden H, Kasher P, Kirby J et al (2007) Microarray analysis of the cellular pathways involved in the adaptation to and progression of motor neuron injury in the SOD1 G93A mouse model of familial ALS. J Neurosci 27:9201–9219
Colucci-Guyon E, Portier MM, Dunia I, Paulin D, Pournin S et al (1994) Mice lacking vimentin develop and reproduce without an obvious phenotype. Cell 79:679–694
Yu H, Kessler J, Shen J (2000) Heterogeneous populations of ES cells in the generation of a floxed Presenilin-1 allele. Genesis 26:5–8
Rogers DC, Fisher EM, Brown SD, Peters J, Hunter AJ et al (1997) Behavioral and functional analysis of mouse phenotype: SHIRPA, a proposed protocol for comprehensive phenotype assessment. Mamm Genome 8:711–713
Masuya H, Inoue M, Wada Y, Shimizu A, Nagano J et al (2005) Implementation of the modified-SHIRPA protocol for screening of dominant phenotypes in a large-scale ENU mutagenesis program. Mamm Genome 16:829–837
Flaherty L (1981) Congenic strains. In: Foster HL, Small JD, Fox JG (eds) The mouse in biomedical research. Academic Press, New York, pp 215–222
Lin DY, Huang CC, Hsieh YT, Lin HC, Pao PC et al (2013) Analysis of the interaction between Zinc finger protein 179 (Znf179) and promyelocytic leukemia zinc finger (Plzf). J Biomed Sci 20:98–107
Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR et al (2014) An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci 34:11929–11947
Jopling HM, Odell AF, Pellet-Many C, Latham AM, Frankel P et al (2014) Endosome-to-plasma membrane recycling of VEGFR2 receptor tyrosine kinase regulates endothelial function and blood vessel formation. Cell 29:363–385
Lee MY, Skoura A, Park EJ, Landskroner-Eiger S, Jozsef L et al (2014) Dynamin 2 regulation of integrin endocytosis, but not VEGF signaling, is crucial for developmental angiogenesis. Development 141:1465–1472
Barkovich AJ, Kuzniecky RI, Jackson GD, Guerrini R, Dobyns WB (2005) A developmental and genetic classification for malformations of cortical development. Neurology 65:1873–1887
Barkovich AJ, Guerrini R, Kuzniecky RI, Jackson GD, Dobyns WB (2012) A developmental and genetic classification for malformations of cortical development: update 2012. Brain 135:1348–1369
Xiang C, Baubet V, Pal S, Holderbaum L, Tatard V et al (2012) RP58/ZNF238 directly modulates proneurogenic gene levels and is required for neuronal differentiation and brain expansion. Cell Death Differ 19:692–702
Boehm SL II, Schafer GL, Phillips TJ, Browman KE, Crabbe JC (2000) ntra-nucleus accumbens shell injections of R(+)- and S(−)-baclofen bidirectionally alter binge-like ethanol, but not saccharin, intake in C57Bl/6J mice. Behav Brain Res 272:238–47
Piot-Grosjean O, Wahl F, Gobbo O, Stutzmann JM (2001) Assessment of sensorimotor and cognitive deficits induced by a moderate traumatic injury in the right parietal cortex of the rat. Neurobiol Dis 8:1082–1093
Stanley JL, Lincoln RJ, Brown TA, McDonald LM, Dawson GR, Reynolds DS (2005) The mouse beam walking assay offers improved sensitivity over the mouse rotarod in determining motor coordination deficits induced by benzodiazepines. J Psychopharmacol 19:221–227
Lømo T (1966) Frequency potentiation of excitatory synaptic activity in the dentate area of the hippocampal formation. Acta Physiol Scand 68:128
Malenka RC (1994) Synaptic plasticity in the hippocampus: LTP and LTD. Cell 78:535–538
Shi SH, Hayashi Y, Petralia RS, Zaman SH, Wenthold RJ et al (1999) Rapid spine delivery and redistribution of AMPA receptors after synaptic NMDA receptor activation. Science 284:1811–1816
Bredt DS, Nicoll RA (2003) AMPA receptor trafficking at excitatory synapses. Neuron 40:361–379
Park M, Penick EC, Edwards JG, Kauer JA, Ehlers MD (2004) Recycling endosomes supply AMPA receptors for LTP. Science 305:1972–1975
Huganir RL, Nicoll RA (2013) AMPARs and synaptic plasticity: the last 25 years. Neuron 80:704–717
Acknowledgments
This work was supported by the National Science Council, Republic of China (NSC 102-2320-B-038-024), Ministry of Science and Technology, Republic of China (MOST 103-2320-B-038-034, MOST 103-2320-B-197-002), and Taipei Medical University (TMU101-AE3-Y24). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We thank the technical services provided by the “Transgenic Mouse Model Core Facility of the National Core Facility Program for Biotechnology, National Science Council” and the “Gene Knockout Mouse Core Laboratory of National Taiwan University Center of Genomic Medicine.” We also thank the technical supports from the Taiwan Mouse Clinic (MOST 104-2325-B-001-011) which is funded by the National Research Program for Biopharmaceuticals (NRPB) at the Ministry of Science and Technology (MOST) of Taiwan. We thank Ms. Christine Chin-jung Hsieh for the English language editing of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Jen-Hui Tsou and Ying-Chen Yang contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Fig. 1
MRI images of mouse brain. Axial (a) and coronal (b–d) views of MRI. Data was obtained from the 5 wild type (Rnf112 +/+) and 4 homozygous Rnf112-knockout (Rnf112 −/−) mice (PDF 16652 kb)
Supplemental Fig. 2
Expression of Rnf112 in umbilical cord. Western blot analysis of Rnf112 and β-actin expressions in the indicated tissues (PDF 838 kb)
Rights and permissions
About this article
Cite this article
Tsou, JH., Yang, YC., Pao, PC. et al. Important Roles of Ring Finger Protein 112 in Embryonic Vascular Development and Brain Functions. Mol Neurobiol 54, 2286–2300 (2017). https://doi.org/10.1007/s12035-016-9812-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-016-9812-7