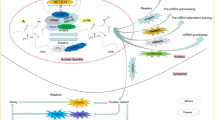
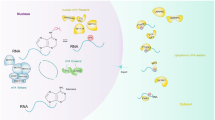
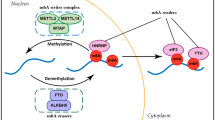
Abstract
RNA methylation, which was identified back in 1970s, has gained remarkable interest in recent years as it was shown to be a reversible modification involved in many cellular processes like mRNA and miRNA processing, mRNA localisation, translation suppression, or activation. These, in turn, affect important bioprocesses such as tissue development, sex determination, and DNA damage response. Important group of proteins are responsible for adding, recognizing, and removing the methyl group to and from the RNA molecules, which are referred as writers, readers, and erasers, respectively. If any of the processes is not strictly controlled, this can cause abnormalities in gene expression, which result in diseases including cancers such as lung, pancreas, glioblastoma, and breast cancer. Mechanisms of RNA methylation and its role in various cancer types and diagnostic methods for RNA methylation are discussed in this article.



Similar content being viewed by others
References
Moore LD, Le T, Fan G. DNA methylation its basic function. Neuropsychopharmacology 2013;38(1):23–38.
Torres IO, Fujimori DG. Functional coupling between writers, erasers and readers of histone and DNA methylation. Curr Opin Struct Biol. 2015;35:68–75.
Curradi M, et al. Molecular mechanisms of gene silencing mediated by DNA methylation. Mol Cell Biol. 2002;22(9):3157–73.
Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet. 2012;13(5):343–57.
EPIGENTEK. RNA methylation analysis made easy. 2010 http://dx.doi.org/10.1093/nar/gkp1117.
Liu J, Jia G. Methylation modifications in eukaryotic messenger RNA. J Genet Genomics. 2014;41(1):21–33.
Roignant JY, Soller M. m(6)A in mRNA: an ancient mechanism for fine-tuning gene expression. Trends Genet. 2017;33(6):380–90.
Deng X, et al. Role of N(6)-methyladenosine modification in cancer. Curr Opin Genet Dev. 2018;48:1–7.
Boccaletto P, et al. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018;46(D1):D303-d307.
Squires JE, et al. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012;40(11):5023–33.
Motorin Y, Lyko F, Helm M. 5-Methylcytosine in RNA: detection, enzymatic formation and biological functions. Nucleic Acids Res. 2010;38(5):1415–30.
Amort T, et al. Distinct 5-methylcytosine profiles in poly(A) RNA from mouse embryonic stem cells and brain. Genome Biol. 2017. 18(1).
Helm M. Post-transcriptional nucleotide modification and alternative folding of RNA. Nucleic Acids Res. 2006;34(2):721–33.
Pan Y, et al. Multiple functions of m6A RNA methylation in cancer. J Hematol Oncol 2018;11(1):48.
Zhou KI, Pan T. Structures of the m6A methyltransferase complex: two subunits with distinct but coordinated roles. Mol Cell. 2016;63(2):183–5.
Ping XL, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24(2):177–89.
Zheng G, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49(1):18–29.
Jia G, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7(12):885–7.
Jia G, Fu Y, He C. Reversible RNA adenosine methylation in biological regulation. Trends Genet. 2013;29(2):108–15.
Wang X, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505(7481):117–20.
Wang X, et al. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161(6):1388–99.
Shi H, et al. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 2017;27(3):315–28.
Meyer KD, et al. 5′ UTR m(6)A promotes cap-independent translation. Cell. 2015;163(4):999–1010.
Alarcon CR, et al. HNRNPA2B1 is a mediator of m(6)A-dependent nuclear RNA processing events. Cell. 2015;162(6):1299–308.
Liu N, et al. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518(7540):560–4.
Jaffrey SR, Kharas MG. Emerging links between m6A and misregulated mRNA methylation in cancer. Genome Med. 2017. 9.
Vu LP, et al. The N(6)-methyladenosine (m(6)A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med. 2017;23(11):1369–76.
Weng H, et al. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m(6)A modification. Cell Stem Cell. 2018;22(2):191–205.e9.
Li Z, et al. FTO plays an oncogenic role in acute myeloid leukemia as a N(6)-methyladenosine RNA demethylase. Cancer Cell. 2017;31(1):127–41.
Mochizuki S, Okada Y. ADAMs in cancer cell proliferation progression. Cancer Sci. 2007;98(5):621–8.
Cui Q, et al. m(6)A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 2017;18(11):2622–34.
Zhang S, et al. m(6)A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 2017;31(4):591–606.e6.
Li Y, Zhang S, Huang S. FoxM1: a potential drug target for glioma. Future Oncol. 2012;8(3):223–6.
Zhang C, et al., Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m(6)A-demethylation of NANOG mRNA. Proc Natl Acad Sci USA 2016. 113(14):E2047-56
Cai X, et al. HBXIP-elevated methyltransferase METTL3 promotes the progression of breast cancer via inhibiting tumor suppressor let-7 g. Cancer Lett. 2018;415:11–9.
Chen M, et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67(6):2254–70.
Chen J, et al. YTH domain family 2 orchestrates epithelial-mesenchymal transition/proliferation dichotomy in pancreatic cancer cells. Cell Cycle. 2017;16(23):2259–71.
Yuan S, et al. Methylation by NSun2 represses the levels and function of microRNA 125b. Mol Cell Biol. 2014;34(19):3630–41.
Pollex T, Hanna K, Schaefer M. Detection of cytosine methylation in RNA using bisulfite sequencing. Cold Spring Harb Protoc 2010. 2010(10): p. pdb.prot5505.
Peer E, Rechavi G, Dominissini D. Epitranscriptomics: regulation of mRNA metabolism through modifications. Curr Opin Chem Biol. 2017;41:93–8.
Dominissini D, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-sEq. Nature. 2012;485(7397):201–6.
Linder B, et al. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods. 2015;12(8):767–72.
Schwartz S, Motorin Y. Next-generation sequencing technologies for detection of modified nucleotides in RNAs. RNA Biol. 2017;14(9):1124–37.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors certify that they have NO affiliations with or involvement in any organization or entity with any financial or non-financial interest in the subject matter or materials discussed in this manuscript.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Tuncel, G., Kalkan, R. Importance of m N6-methyladenosine (m6A) RNA modification in cancer. Med Oncol 36, 36 (2019). https://doi.org/10.1007/s12032-019-1260-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-019-1260-6