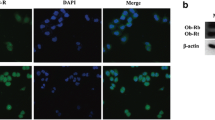
Abstract
In postmenopausal women, adipositas represents a serious risk factor for cancer development and progression. White adipose tissue secretes the 16 kDa hormone leptin which plays a key role in the regulation of appetite and metabolism. An increasing number of reports indicate that leptin also interferes with signal transduction pathways implicated in the development of breast cancer. In our previous study, we identified the estrogen receptor alpha (ERα) as a relevant enhancer of leptin-induced signal transduction leading to transactivation of signal transducer and activator of transcription 3 (Stat3). The purpose of this study is the investigation of specific target gene expression in response to leptin-mediated Stat3 signaling. We performed a comprehensive microarray analysis of ERα-positive and ERα-negative MDA-MB-231 cells upon leptin treatment and identified 49 genes which showed a significant ERα-dependent regulation in leptin-treated MDA-MB-231 cells. There was no intersection with genes which were merely up- or downregulated by ERα expression and only 9 and 11 genes overlapping targets which were regulated by leptin stimulation either in ERα-expressing or ERα-negative MDA-MB-231 cells, respectively. To demonstrate the specificity, expression of three target genes was validated by quantitative real-time PCR. In conclusion, these data imply that leptin can induce a different set of target genes dependent on ERα expression, which might contribute to the development and progression of cancer diseases.



Similar content being viewed by others
Abbreviations
- ERα:
-
Estrogen receptor alpha
- Ob-RL:
-
Leptin receptor
- Stat3:
-
Signal transducer and activator of transcription 3
References
G.L. Anderson, M.L. Neuhouser, Obesity and the risk for premenopausal and postmenopausal breast cancer. Cancer Prev. Res. (Phila.) 5, 515–521 (2012)
A. Ray, M.P. Cleary, Obesity and breast cancer: a clinical biochemistry perspective. Clin. Biochem. 45, 189–197 (2012)
C. Garofalo, E. Surmacz, Leptin and cancer. J. Cell. Physiol. 207, 12–22 (2006)
C. Deglise, C. Bouchardy, M. Burri, M. Usel, I. Neyroud-Caspar, G. Vlastos, P.O. Chappuis, M. Ceschi, S. Ess, M. Castiglione, E. Rapiti, H.M. Verkooijen, Impact of obesity on diagnosis and treatment of breast cancer. Breast Cancer Res. Treat. 120, 185–193 (2010)
B. Majed, A. Dozol, L. Ribassin-Majed, K. Senouci, B. Asselain, Increased risk of contralateral breast cancers among overweight and obese women: a time-dependent association. Breast Cancer Res. Treat. 126, 729–738 (2011)
G. Fruhbeck, Intracellular signalling pathways activated by leptin. Biochem. J. 393, 7–20 (2006)
K. Lang, J. Ratke, Leptin and adiponectin: new players in the field of tumor cell and leukocyte migration. Cell Commun. Signal. 7, 27 (2009)
H. Feng, L. Zheng, Z. Feng, Y. Zhao, N. Zhang, The role of leptin in obesity and the potential for leptin replacement therapy. Endocrine (2012)
N. Ghilardi, S. Ziegler, A. Wiestner, R. Stoffel, M.H. Heim, R.C. Skoda, Defective STAT signaling by the leptin receptor in diabetic mice. Proc. Natl. Acad. Sci. U.S.A. 93, 6231–6235 (1996)
C. Bjorbaek, S. Uotani, B. da Silva, J.S. Flier, Divergent signaling capacities of the long and short isoforms of the leptin receptor. J. Biol. Chem. 272, 32686–32695 (1997)
D. Cirillo, A.M. Rachiglio, R. la Montagna, A. Giordano, N. Normanno, Leptin signaling in breast cancer: an overview. J. Cell. Biochem. 105, 956–964 (2008)
N. Kiuchi, K. Nakajima, M. Ichiba, T. Fukada, M. Narimatsu, K. Mizuno, M. Hibi, T. Hirano, STAT3 is required for the gp130-mediated full activation of the c-myc gene. J. Exp. Med. 189, 63–73 (1999)
D. Sinibaldi, W. Wharton, J. Turkson, T. Bowman, W.J. Pledger, R. Jove, Induction of p21WAF1/CIP1 and cyclin D1 expression by the Src oncoprotein in mouse fibroblasts: role of activated STAT3 signaling. Oncogene 19, 5419–5427 (2000)
J.V. Alvarez, P.G. Febbo, S. Ramaswamy, M. Loda, A. Richardson, D.A. Frank, Identification of a genetic signature of activated signal transducer and activator of transcription 3 in human tumors. Cancer Res. 65, 5054–5062 (2005)
L. Bjornstrom, M. Sjoberg, Signal transducers and activators of transcription as downstream targets of nongenomic estrogen receptor actions. Mol. Endocrinol. 16, 2202–2214 (2002)
L. Canesi, C. Ciacci, M. Betti, L.C. Lorusso, B. Marchi, S. Burattini, E. Falcieri, G. Gallo, Rapid effects of 17beta-estradiol on cell signaling and function of Mytilus hemocytes. Gen. Comp. Endocrinol. 136, 58–71 (2004)
S. Catalano, L. Mauro, S. Marsico, C. Giordano, P. Rizza, V. Rago, D. Montanaro, M. Maggiolini, M.L. Panno, S. Ando, Leptin induces, via ERK1/ERK2 signal, functional activation of estrogen receptor alpha in MCF-7 cells. J. Biol. Chem. 279, 19908–19915 (2004)
P. Ciana, S. Ghisletti, P. Mussi, I. Eberini, E. Vegeto, A. Maggi, Estrogen receptor alpha, a molecular switch converting transforming growth factor-alpha-mediated proliferation into differentiation in neuroblastoma cells. J. Biol. Chem. 278, 31737–31744 (2003)
N.A. Binai, A. Damert, G. Carra, S. Steckelbroeck, J. Lower, R. Lower, S. Wessler, Expression of estrogen receptor alpha increases leptin-induced STAT3 activity in breast cancer cells. Int. J. Cancer 127, 55–66 (2010)
S. Wessler, C. Otto, N. Wilck, V. Stangl, K.H. Fritzemeier, Identification of estrogen receptor ligands leading to activation of non-genomic signaling pathways while exhibiting only weak transcriptional activity. J. Steroid Biochem. Mol. Biol. 98, 25–35 (2006)
L. Tora, A. Mullick, D. Metzger, M. Ponglikitmongkol, I. Park, P. Chambon, The cloned human oestrogen receptor contains a mutation which alters its hormone binding properties. EMBO J. 8, 1981–1986 (1989)
R.A. Irizarry, B. Hobbs, F. Collin, Y.D. Beazer-Barclay, K.J. Antonellis, U. Scherf, T.P. Speed, Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264 (2003)
J. Oliveros, VENNY. An interactive tool for comparing lists with Venn diagrams. http://bioinfogp.cnb.csic.es/tools/venny/. Accessed Aug 2012
T.D. Schmittgen, K.J. Livak, Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108 (2008)
K. Subik, J.F. Lee, L. Baxter, T. Strzepek, D. Costello, P. Crowley, L. Xing, M.C. Hung, T. Bonfiglio, D.G. Hicks, P. Tang, The expression patterns of ER, PR, HER2, CK5/6, EGFR, Ki-67 and AR by immunohistochemical analysis in breast cancer cell lines. Breast cancer 4, 35–41 (2010)
R. Fusco, M. Galgani, C. Procaccini, R. Franco, G. Pirozzi, L. Fucci, P. Laccetti, G. Matarese, Cellular and molecular crosstalk between leptin receptor and estrogen receptor-{alpha} in breast cancer: molecular basis for a novel therapeutic setting. Endocr. Relat. Cancer 17, 373–382 (2010)
C. Meazza, G. Vitale, S. Pagani, D. Castaldi, G. Ogliari, D. Mari, K. Laarej, C. Tinelli, M. Bozzola, Common adipokine features of neonates and centenarians. J. Pediatr. Endocrinol. Metab. 24, 953–957 (2011)
R. Ostan, L. Bucci, E. Cevenini, M. G. Palmas, E. Pini, M. Scurti, R. Vescovini, C. Caruso, D. Mari, G. Vitale, C. Franceschi, D. Monti, Metabolic syndrome in the offspring of centenarians: focus on prevalence, components, and adipokines. Age (2012). doi:10.1007/s11357-012-9483-x
A. Ptak, E. Kolaczkowska, E. L. Gregoraszczuk, Leptin stimulation of cell cycle and inhibition of apoptosis gene and protein expression in OVCAR-3 ovarian cancer cells. Endocrine (2012). doi:10.1007/s12020-012-9788-7
P. Hekerman, J. Zeidler, S. Korfmacher, S. Bamberg-Lemper, H. Knobelspies, L. Zabeau, J. Tavernier, W. Becker, Leptin induces inflammation-related genes in RINm5F insulinoma cells. BMC Mol. Biol. 8, 41 (2007)
S. Loffreda, S.Q. Yang, H.Z. Lin, C.L. Karp, M.L. Brengman, D.J. Wang, A.S. Klein, G.B. Bulkley, C. Bao, P.W. Noble, M.D. Lane, A.M. Diehl, Leptin regulates proinflammatory immune responses. FASEB J. 12, 57–65 (1998)
D.J. Dauer, B. Ferraro, L. Song, B. Yu, L. Mora, R. Buettner, S. Enkemann, R. Jove, E.B. Haura, Stat3 regulates genes common to both wound healing and cancer. Oncogene 24, 3397–3408 (2005)
T. Jarde, F. Caldefie-Chezet, N. Goncalves-Mendes, F. Mishellany, C. Buechler, F. Penault-Llorca, M.P. Vasson, Involvement of adiponectin and leptin in breast cancer: clinical and in vitro studies. Endocr. Relat. Cancer 16, 1197–1210 (2009)
C.N. Perera, H.G. Chin, N. Duru, I.G. Camarillo, Leptin-regulated gene expression in MCF-7 breast cancer cells: mechanistic insights into leptin-regulated mammary tumor growth and progression. J. Endocrinol. 199, 221–233 (2008)
B. De Cat, G. David, Developmental roles of the glypicans. Semin. Cell Dev. Biol. 12, 117–125 (2001)
C. Zhang, S. Zhang, D. Zhang, Z. Zhang, Y. Xu, S. Liu, A lung cancer gene GPC5 could also be crucial in breast cancer. Mol. Genet. Metab. 103, 104–105 (2011)
P. Hulpiau, F. van Roy, Molecular evolution of the cadherin superfamily. Int. J. Biochem. Cell Biol. 41, 349–369 (2009)
M. Miyagawa, S.Y. Nishio, S. Usami, Prevalence and clinical features of hearing loss patients with CDH23 mutations: a large cohort study. PLoS ONE 7, e40366 (2012)
M. Apostolopoulou, L. Ligon, Cadherin-23 mediates heterotypic cell–cell adhesion between breast cancer epithelial cells and fibroblasts. PLoS ONE 7, e33289 (2012)
C.T. Dolphin, E.A. Shephard, S. Povey, R.L. Smith, I.R. Phillips, Cloning, primary sequence and chromosomal localization of human FMO2, a new member of the flavin-containing mono-oxygenase family. Biochem. J. 287, 261–267 (1992)
F. Fialka, R.M. Gruber, R. Hitt, L. Opitz, E. Brunner, H. Schliephake, F.J. Kramer, CPA6, FMO2, LGI1, SIAT1 and TNC are differentially expressed in early- and late-stage oral squamous cell carcinoma—a pilot study. Oral Oncol. 44, 941–948 (2008)
S.K. Krueger, J.E. Vandyke, D.E. Williams, R.N. Hines, The role of flavin-containing monooxygenase (FMO) in the metabolism of tamoxifen and other tertiary amines. Drug Metab. Rev. 38, 139–147 (2006)
C. Mani, D. Kupfer, Cytochrome P-450-mediated activation and irreversible binding of the antiestrogen tamoxifen to proteins in rat and human liver: possible involvement of flavin-containing monooxygenases in tamoxifen activation. Cancer Res. 51, 6052–6058 (1991)
N.K. Verma, J. Dourlat, A.M. Davies, A. Long, W.Q. Liu, C. Garbay, D. Kelleher, Y. Volkov, STAT3–stathmin interactions control microtubule dynamics in migrating T-cells. J. Biol. Chem. 284, 12349–12362 (2009)
J. Azare, K. Leslie, H. Al-Ahmadie, W. Gerald, P.H. Weinreb, S.M. Violette, J. Bromberg, Constitutively activated Stat3 induces tumorigenesis and enhances cell motility of prostate epithelial cells through integrin beta 6. Mol. Cell. Biol. 27, 4444–4453 (2007)
D.C. Ng, B.H. Lin, C.P. Lim, G. Huang, T. Zhang, V. Poli, X. Cao, Stat3 regulates microtubules by antagonizing the depolymerization activity of stathmin. J. Cell Biol. 172, 245–257 (2006)
S.R. Walker, M. Chaudhury, D.A. Frank, STAT3 Inhibition by microtubule-targeted drugs: dual molecular effects of chemotherapeutic agents. Mol. Cell. Pharmacol. 3, 13–19 (2011)
Acknowledgments
We thank Dr. Bernd Groner (Georg Speyer Haus, Frankfurt, Germany) for providing MDA-MB-231 cells.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Binai, N.A., Carra, G., Löwer, J. et al. Differential gene expression in ERα-positive and ERα-negative breast cancer cells upon leptin stimulation. Endocrine 44, 496–503 (2013). https://doi.org/10.1007/s12020-013-9897-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12020-013-9897-y