Abstract
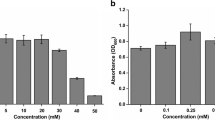
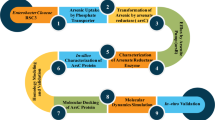
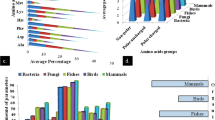
The arsC gene–encoded arsenate reductase is a vital catalytic enzyme for remediation of environmental arsenic (As). Microorganisms containing the arsC gene can convert pentavalent arsenate (As[V]) to trivalent arsenite (As[III]) to be either retained in the bacterial cell or released into the air. The molecular mechanism governing this process is unknown. Here we present an in silico model of the enzyme to describe their probable active site cavities using SCFBio servers. We retrieved the amino acid sequence of bacterial arsenate reductase enzymes in FASTA format from the NCBI database. Enzyme structure was predicted using the I-TASSER server and visualized using PyMOL tools. The ProSA and the PROCHECK servers were used to evaluate the overall significance of the predicted model. Accordingly, arsenate reductase from Streptococcus pyogenes, Oligotropha carboxidovorans OM5, Rhodopirellula baltica SH 1, and Serratia ureilytica had the highest quality scores with statistical significance. The plausible cavities of the active site were identified in our examined arsenate reductase enzymes which were abundant in glutamate and lysine residues with 6 to 16 amino acids. This in silico experiment may contribute greatly to the remediation of arsenic pollution through the utilization of microbial species.






Similar content being viewed by others
References
Rahman, S., Kim, K.-H., Saha, S. K., Swaraz, A., & Paul, D. K. (2014). Review of remediation techniques for arsenic (As) contamination: a novel approach utilizing bio-organisms. Journal of Environmental Management, 134, 175–185.
Liu, Z., Li, W., Qi, H., Song, G., Ding, D., Fu, Z., Liu, J., & Tang, J. (2012). Arsenic accumulation and distribution in the tissues of inbred lines in maize (Zea mays L.). Genetic Resources and Crop Evolution, 59(8), 1705–1711.
Rahman, M. S., Biswas, P. K., Al Hasan, S. M., Rahman, M. M., Lee, S. H., Kim, K.-H., Rahman, S. M., & Islam, M. R. (2018). The occurrences of heavy metals in farmland soils and their propagation into paddy plants. Environmental Monitoring and Assessment, 190(4), 201.
Mandal, B. K., & Suzuki, K. T. (2002). Arsenic round the world: a review. Talanta, 58(1), 201–235.
Düring, R.-A., Hoß, T., & Gäth, S. (2003). Sorption and bioavailability of heavy metals in long-term differently tilled soils amended with organic wastes. Science of the Total Environment, 313(1–3), 227–234.
Tamaki, S., & Frankenberger, W. T. (1992). Environmental biochemistry of arsenic. In Reviews of environmental contamination and toxicology (pp. 79–110). Berlin: Springer.
Cervantes, C., Ji, G., Ramírez, J. L., & Silver, S. (1994). Resistance to arsenic compounds in microorganisms. FEMS Microbiology Reviews, 15(4), 355–367.
Muller, D., Lievremont, D., Simeonova, D. D., Hubert, J.-C., & Lett, M.-C. (2003). Arsenite oxidase aox genes from a metal-resistant β-proteobacterium. Journal of Bacteriology, 185(1), 135–141.
Huber, R., Sacher, M., Vollmann, A., Huber, H., & Rose, D. (2000). Respiration of arsenate and selenate by hyperthermophilic archaea. Systematic and Applied Microbiology, 23(3), 305–314.
Rosen, B. P. (2002). Biochemistry of arsenic detoxification. FEBS Letters, 529(1), 86–92.
Martin, P., DeMel, S., Shi, J., Gladysheva, T., Gatti, D. L., Rosen, B. P., & Edwards, B. F. (2001). Insights into the structure, solvation, and mechanism of ArsC arsenate reductase, a novel arsenic detoxification enzyme. Structure, 9(11), 1071–1081.
Ji, G., Garber, E. A., Armes, L. G., Chen, C.-M., Fuchs, J. A., & Silver, S. (1994). Arsenate reductase of Staphylococcus aureus plasmid pI258. Biochemistry, 33(23), 7294–7299.
Mukhopadhyay, R., Shi, J., & Rosen, B. P. (2000). Purification and characterization of Acr2p, the Saccharomyces cerevisiae arsenate reductase. Journal of Biological Chemistry, 275(28), 21149–21157.
Jeanmougin, F., Thompson, J. D., Gouy, M., Higgins, D. G., & Gibson, T. J. (1998). Multiple sequence alignment with Clustal X. Trends in Biochemical Sciences, 23(10), 403–405.
Kumar, S., Stecher, G., Li, M., Knyaz, C., & Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution, 35(6), 1547–1549.
Roy, A., Kucukural, A., & Zhang, Y. (2010). I-TASSER: a unified platform for automated protein structure and function prediction. Nature Protocols, 5(4), 725–738.
Yang, J., Yan, R., Roy, A., Xu, D., Poisson, J., & Zhang, Y. (2015). The I-TASSER suite: protein structure and function prediction. Nature Methods, 12(1), 7–8.
Yang, J., & Zhang, Y. (2015). I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Research, 43(W1), W174–W181.
Laskowski, R. A., Jabłońska, J., Pravda, L., Vařeková, R. S., & Thornton, J. M. (2018). PDBsum: structural summaries of PDB entries. Protein Science, 27(1), 129–134.
Ramachandran, G. N. (1963). Stereochemistry of polypeptide chain configurations. Journal of Molecular Biology, 7(1), 95–99.
Laskowski, R. A., Rullmann, J. A. C., MacArthur, M. W., Kaptein, R., & Thornton, J. M. (1996). AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. Journal of Biomolecular NMR, 8(4), 477–486.
Wiederstein, M., & Sippl, M. J. (2007). ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Research, 35(suppl_2), W407–W410.
Eisenberg, D., Lüthy, R., & Bowie, J. U. (1997). [20] VERIFY3D: assessment of protein models with three-dimensional profiles. In Methods in enzymology (Vol. 277, pp. 396–404). Cambridge: Academic Press.
Colovos, C., & Yeates, T. O. (1993). Verification of protein structures: patterns of nonbonded atomic interactions. Protein Science, 2(9), 1511–1519.
Gasteiger, E., Hoogland, C., Gattiker, A., Wilkins, M. R., Appel, R. D., & Bairoch, A. (2005). Protein identification and analysis tools on the ExPASy server. In The proteomics protocols handbook (pp. 571–607). Berlin: Springer.
Singh, T., Biswas, D., & Jayaram, B. (2011). AADS-an automated active site identification, docking, and scoring protocol for protein targets based on physicochemical descriptors. Journal of Chemical Information and Modeling, 51(10), 2515–2527.
Martínez-Rosell, G., Giorgino, T., & De Fabritiis, G. (2017). PlayMolecule ProteinPrepare: a web application for protein preparation for molecular dynamics simulations. Journal of Chemical Information and Modeling, 57(7), 1511–1516.
Doerr, S., Harvey, M. J., Noé, F., & De Fabritiis, G. (2016). HTMD: high-throughput molecular dynamics for molecular discovery. Journal of Chemical Theory and Computation, 12(4), 1845–1852.
Govarthanan, M., Lee, S.-M., Kamala-Kannan, S., & Oh, B.-T. (2015). Characterization, real-time quantification and in silico modeling of arsenate reductase (arsC) genes in arsenic-resistant Herbaspirillum sp. GW103. Research in Microbiology, 166(3), 196–204.
Vyas, V. K., Ukawala, R. D., Ghate, M., & Chintha, C. (2012). Homology modeling a fast tool for drug discovery: current perspectives. Indian Journal of Pharmaceutical Sciences, 74(1), 1–17.
DeMel, S., Shi, J., Martin, P., Rosen, B. P., & Edwards, B. F. P. (2004). Arginine 60 in the ArsC arsenate reductase of E. coli plasmid R773 determines the chemical nature of the bound As(III) product. Protein Science, 13(9), 2330–2340.
Roos, G., Buts, L., Van Belle, K., Brosens, E., Geerlings, P., Loris, R., Wyns, L., & Messens, J. (2006). Interplay between ion binding and catalysis in the thioredoxin-coupled arsenate reductase family. Journal of Molecular Biology, 360(4), 826–838.
Barman, U. D., Saha, S. K., Kader, M. A., Jamal, M. A. H. M., Sharma, S. P., Samad, A., & Rahman, M. S. (2020). Clinicopathological and prognostic significance of GPC3 in human breast cancer and its 3D structure prediction. Network Modeling Analysis in Health Informatics and Bioinformatics, 9, 1–18.
Nain, Z., Sayed, S. B., Karim, M. M., Islam, M. A., & Adhikari, U. K. (2019). Energy-optimized pharmacophore coupled virtual screening in the discovery of quorum sensing inhibitors of LasR protein of Pseudomonas aeruginosa. Journal of Biomolecular Structure and Dynamics. https://doi.org/10.1080/07391102.2019.1700168.
Funding
This research was supported by a grant from the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning (Grant No: 2016R1E1A1A01940995).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict interest.
Research Involving Human Participants and/or Animals
No human and animal participants are involved.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Rahman, M.S., Hossain, M.S., Saha, S.K. et al. Homology Modeling and Probable Active Site Cavity Prediction of Uncharacterized Arsenate Reductase in Bacterial spp.. Appl Biochem Biotechnol 193, 1–18 (2021). https://doi.org/10.1007/s12010-020-03392-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-020-03392-w