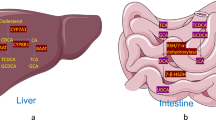
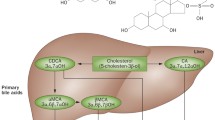
Abstract
Purpose of Review
Herein, we review the role of FXR and TGR5 in the regulation of hepatic bile acid metabolism, with a focus on how our understanding of bile acid metabolic regulation by these receptors has evolved in recent years and how this improved understanding may facilitate targeting bile acids for type 2 diabetes treatment.
Recent Findings
Bile acid profile is a key regulator of metabolic homeostasis. Inhibition of expression of the enzyme that is required for cholic acid synthesis and thus determines bile acid profile, Cyp8b1, may be an effective target for type 2 diabetes treatment. FXR and, more recently, TGR5 have been shown to regulate bile acid metabolism and Cyp8b1 expression and, therefore, may provide a mechanism with which to target bile acid profile for type 2 diabetes treatment.
Summary
Inhibition of Cyp8b1 expression is a promising therapeutic modality for type 2 diabetes; however, further work is needed to fully understand the pathways regulating Cyp8b1 expression.


Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Lefebvre P, Cariou B, Lien F, Kuipers F, Staels B. Role of bile acids and bile acid receptors in metabolic regulation. Physiol Rev. 2009;89(1):147–91. https://doi.org/10.1152/physrev.00010.2008.
Thomas C, Pellicciari R, Pruzanski M, Auwerx J, Schoonjans K. Targeting bile-acid signalling for metabolic diseases. Nat Rev Drug Discov. 2008;7(8):678–93. https://doi.org/10.1038/nrd2619.
Haeusler RA, Astiarraga B, Camastra S, Accili D, Ferrannini E. Human insulin resistance is associated with increased plasma levels of 12alpha-hydroxylated bile acids. Diabetes. 2013;62(12):4184–91. https://doi.org/10.2337/db13-0639.
Heuman DM. Quantitative estimation of the hydrophilic-hydrophobic balance of mixed bile salt solutions. J Lipid Res. 1989;30(5):719–30.
Mahmoud AA, Elshazly SM. Ursodeoxycholic acid ameliorates fructose-induced metabolic syndrome in rats. PLoS One. 2014;9(9):e106993. https://doi.org/10.1371/journal.pone.0106993.
Ozcan U, Yilmaz E, Ozcan L, Furuhashi M, Vaillancourt E, Smith RO, et al. Chemical chaperones reduce ER stress and restore glucose homeostasis in a mouse model of type 2 diabetes. Science. 2006;313(5790):1137–40. https://doi.org/10.1126/science.1128294.
Shima KR, Ota T, Kato KI, Takeshita Y, Misu H, Kaneko S, et al. Ursodeoxycholic acid potentiates dipeptidyl peptidase-4 inhibitor sitagliptin by enhancing glucagon-like peptide-1 secretion in patients with type 2 diabetes and chronic liver disease: a pilot randomized controlled and add-on study. BMJ Open Diabetes Res Care. 2018;6(1):e000469. https://doi.org/10.1136/bmjdrc-2017-000469.
Cariou B, Chetiveaux M, Zair Y, Pouteau E, Disse E, Guyomarc'h-Delasalle B, et al. Fasting plasma chenodeoxycholic acid and cholic acid concentrations are inversely correlated with insulin sensitivity in adults. Nutr Metab (Lond). 2011;8(1):48. https://doi.org/10.1186/1743-7075-8-48.
Chung SJ, Lee CH, Lee HS, Kim ST, Sohn UD, Park ES, et al. The role of phosphatidylcholine and deoxycholic acid in inflammation. Life Sci. 2014;108(2):88–93. https://doi.org/10.1016/j.lfs.2014.05.013.
Zaborska KE, Lee SA, Garribay D, Cha E, Cummings BP. Deoxycholic acid supplementation impairs glucose homeostasis in mice. PLoS One. 2018;13(7):e0200908. https://doi.org/10.1371/journal.pone.0200908.
Bertaggia E, Jensen KK, Castro-Perez J, Xu Y, Di Paolo G, Chan RB, et al. Cyp8b1 ablation prevents Western diet-induced weight gain and hepatic steatosis because of impaired fat absorption. Am J Physiol Endocrinol Metab. 2017;313(2):E121–33. https://doi.org/10.1152/ajpendo.00409.2016.
Li-Hawkins J, Gafvels M, Olin M, Lund EG, Andersson U, Schuster G, et al. Cholic acid mediates negative feedback regulation of bile acid synthesis in mice. J Clin Invest. 2002;110(8):1191–200. https://doi.org/10.1172/JCI16309.
Murphy C, Parini P, Wang J, Bjorkhem I, Eggertsen G, Gafvels M. Cholic acid as key regulator of cholesterol synthesis, intestinal absorption and hepatic storage in mice. Biochim Biophys Acta. 2005;1735(3):167–75. https://doi.org/10.1016/j.bbalip.2005.06.001.
Slatis K, Gafvels M, Kannisto K, Ovchinnikova O, Paulsson-Berne G, Parini P, et al. Abolished synthesis of cholic acid reduces atherosclerotic development in apolipoprotein E knockout mice. J Lipid Res. 2010;51(11):3289–98. https://doi.org/10.1194/jlr.M009308.
Wang J, Gafvels M, Rudling M, Murphy C, Bjorkhem I, Einarsson C, et al. Critical role of cholic acid for development of hypercholesterolemia and gallstones in diabetic mice. Biochem Biophys Res Commun. 2006;342(4):1382–8. https://doi.org/10.1016/j.bbrc.2006.02.108.
• McGavigan AK, Garibay D, Henseler ZM, Chen J, Bettaieb A, Haj FG, et al. TGR5 contributes to glucoregulatory improvements after vertical sleeve gastrectomy in mice. Gut. 2017;66(2):226–34. https://doi.org/10.1136/gutjnl-2015-309871 This study identifies a role for TGR5 in the regulation of hepatic CYP8B1 expression.
• Pathak P, Liu H, Boehme S, Xie C, Krausz KW, Gonzalez F, et al. Farnesoid X receptor induces Takeda G-protein receptor 5 cross-talk to regulate bile acid synthesis and hepatic metabolism. J Biol Chem. 2017;292(26):11055–69. https://doi.org/10.1074/jbc.M117.784322 This study provides the first description of an interation between TGR5 and FXR.
Pean N, Doignon I, Garcin I, Besnard A, Julien B, Liu B, et al. The receptor TGR5 protects the liver from bile acid overload during liver regeneration in mice. Hepatology. 2013;58(4):1451–60. https://doi.org/10.1002/hep.26463.
Russell DW. The enzymes, regulation, and genetics of bile acid synthesis. Annu Rev Biochem. 2003;72:137–74. https://doi.org/10.1146/annurev.biochem.72.121801.161712.
He D, Barnes S, Falany CN. Rat liver bile acid CoA:amino acid N-acyltransferase: expression, characterization, and peroxisomal localization. J Lipid Res. 2003;44(12):2242–9. https://doi.org/10.1194/jlr.M300128-JLR200.
Ridlon JM, Kang DJ, Hylemon PB, Bajaj JS. Bile acids and the gut microbiome. Curr Opin Gastroenterol. 2014;30(3):332–8. https://doi.org/10.1097/MOG.0000000000000057.
Zhang Y, Klaassen CD. Effects of feeding bile acids and a bile acid sequestrant on hepatic bile acid composition in mice. J Lipid Res. 2010;51(11):3230–42. https://doi.org/10.1194/jlr.M007641.
Hagey LR, Crombie DL, Espinosa E, Carey MC, Igimi H, Hofmann AF. Ursodeoxycholic acid in the Ursidae: biliary bile acids of bears, pandas, and related carnivores. J Lipid Res. 1993;34(11):1911–7.
Bachrach WH, Hofmann AF. Ursodeoxycholic acid in the treatment of cholesterol cholelithiasis. part I. Dig Dis Sci. 1982;27(8):737–61.
Fedorowski T, Salen G, Tint GS, Mosbach E. Transformation of chenodeoxycholic acid and ursodeoxycholic acid by human intestinal bacteria. Gastroenterology. 1979;77(5):1068–73.
Ferdinandusse S, Houten SM. Peroxisomes and bile acid biosynthesis. Biochim Biophys Acta. 2006;1763(12):1427–40. https://doi.org/10.1016/j.bbamcr.2006.09.001.
Chiang JY. Regulation of bile acid synthesis. Front Biosci. 1998;3:d176–93.
Norlin M, Wikvall K. Enzymes in the conversion of cholesterol into bile acids. Curr Mol Med. 2007;7(2):199–218.
Wu Z, Martin KO, Javitt NB, Chiang JY. Structure and functions of human oxysterol 7alpha-hydroxylase cDNAs and gene CYP7B1. J Lipid Res. 1999;40(12):2195–203.
Andersson S, Davis DL, Dahlback H, Jornvall H, Russell DW. Cloning, structure, and expression of the mitochondrial cytochrome P-450 sterol 26-hydroxylase, a bile acid biosynthetic enzyme. J Biol Chem. 1989;264(14):8222–9.
Preuss I, Ludwig MG, Baumgarten B, Bassilana F, Gessier F, Seuwen K, et al. Transcriptional regulation and functional characterization of the oxysterol/EBI2 system in primary human macrophages. Biochem Biophys Res Commun. 2014;446(3):663–8. https://doi.org/10.1016/j.bbrc.2014.01.069.
Reiss AB, Martin KO, Rojer DE, Iyer S, Grossi EA, Galloway AC, et al. Sterol 27-hydroxylase: expression in human arterial endothelium. J Lipid Res. 1997;38(6):1254–60.
Schwarz M, Lund EG, Lathe R, Bjorkhem I, Russell DW. Identification and characterization of a mouse oxysterol 7alpha-hydroxylase cDNA. J Biol Chem. 1997;272(38):23995–4001..
Shanahan CM, Carpenter KL, Cary NR. A potential role for sterol 27-hydroxylase in atherogenesis. Atherosclerosis. 2001;154(2):269–76.
Forman BM, Goode E, Chen J, Oro AE, Bradley DJ, Perlmann T, et al. Identification of a nuclear receptor that is activated by farnesol metabolites. Cell. 1995;81(5):687–93.
Cipriani S, Mencarelli A, Palladino G, Fiorucci S. FXR activation reverses insulin resistance and lipid abnormalities and protects against liver steatosis in Zucker (fa/fa) obese rats. J Lipid Res. 2010;51(4):771–84. https://doi.org/10.1194/jlr.M001602.
De Gottardi A, Touri F, Maurer CA, Perez A, Maurhofer O, Ventre G, et al. The bile acid nuclear receptor FXR and the bile acid binding protein IBABP are differently expressed in colon cancer. Dig Dis Sci. 2004;49(6):982–9.
Raybould HE. Gut microbiota, epithelial function and derangements in obesity. J Physiol. 2012;590(3):441–6. https://doi.org/10.1113/jphysiol.2011.222133.
Makishima M, Okamoto AY, Repa JJ, Tu H, Learned RM, Luk A, et al. Identification of a nuclear receptor for bile acids. Science. 1999;284(5418):1362–5.
Wang H, Chen J, Hollister K, Sowers LC, Forman BM. Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol Cell. 1999;3(5):543–53.
Li Y, Jadhav K, Zhang Y. Bile acid receptors in non-alcoholic fatty liver disease. Biochem Pharmacol. 2013;86(11):1517–24. https://doi.org/10.1016/j.bcp.2013.08.015.
Li F, Jiang C, Krausz KW, Li Y, Albert I, Hao H, et al. Microbiome remodelling leads to inhibition of intestinal farnesoid X receptor signalling and decreased obesity. Nat Commun. 2013;4:2384. https://doi.org/10.1038/ncomms3384.
Parseus A, Sommer N, Sommer F, Caesar R, Molinaro A, Stahlman M, et al. Microbiota-induced obesity requires farnesoid X receptor. Gut. 2017;66(3):429–37. https://doi.org/10.1136/gutjnl-2015-310283.
Sayin SI, Wahlstrom A, Felin J, Jantti S, Marschall HU, Bamberg K, et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17(2):225–35. https://doi.org/10.1016/j.cmet.2013.01.003.
Mueller M, Thorell A, Claudel T, Jha P, Koefeler H, Lackner C, et al. Ursodeoxycholic acid exerts farnesoid X receptor-antagonistic effects on bile acid and lipid metabolism in morbid obesity. J Hepatol. 2015;62(6):1398–404. https://doi.org/10.1016/j.jhep.2014.12.034.
Cariou B, van Harmelen K, Duran-Sandoval D, van Dijk TH, Grefhorst A, Abdelkarim M, et al. The farnesoid X receptor modulates adiposity and peripheral insulin sensitivity in mice. J Biol Chem. 2006;281(16):11039–49. https://doi.org/10.1074/jbc.M510258200.
Fiorucci S, Antonelli E, Rizzo G, Renga B, Mencarelli A, Riccardi L, et al. The nuclear receptor SHP mediates inhibition of hepatic stellate cells by FXR and protects against liver fibrosis. Gastroenterology. 2004;127(5):1497–512.
Fiorucci S, Rizzo G, Donini A, Distrutti E, Santucci L. Targeting farnesoid X receptor for liver and metabolic disorders. Trends Mol Med. 2007;13(7):298–309. https://doi.org/10.1016/j.molmed.2007.06.001.
Mencarelli A, Renga B, D'Amore C, Santorelli C, Graziosi L, Bruno A, et al. Dissociation of intestinal and hepatic activities of FXR and LXRalpha supports metabolic effects of terminal ileum interposition in rodents. Diabetes. 2013;62(10):3384–93. https://doi.org/10.2337/db13-0299.
Swanson HI, Wada T, Xie W, Renga B, Zampella A, Distrutti E, et al. Role of nuclear receptors in lipid dysfunction and obesity-related diseases. Drug Metab Dispos. 2013;41(1):1–11. https://doi.org/10.1124/dmd.112.048694.
Zhang Y, Lee FY, Barrera G, Lee H, Vales C, Gonzalez FJ, et al. Activation of the nuclear receptor FXR improves hyperglycemia and hyperlipidemia in diabetic mice. Proc Natl Acad Sci U S A. 2006;103(4):1006–11. https://doi.org/10.1073/pnas.0506982103.
Dufer M, Horth K, Wagner R, Schittenhelm B, Prowald S, Wagner TF, et al. Bile acids acutely stimulate insulin secretion of mouse beta-cells via farnesoid X receptor activation and K(ATP) channel inhibition. Diabetes. 2012;61(6):1479–89. https://doi.org/10.2337/db11-0815.
Potthoff MJ, Boney-Montoya J, Choi M, He T, Sunny NE, Satapati S, et al. FGF15/19 regulates hepatic glucose metabolism by inhibiting the CREB-PGC-1alpha pathway. Cell Metab. 2011;13(6):729–38. https://doi.org/10.1016/j.cmet.2011.03.019.
Jiang C, Xie C, Li F, Zhang L, Nichols RG, Krausz KW, et al. Intestinal farnesoid X receptor signaling promotes nonalcoholic fatty liver disease. J Clin Invest. 2015;125(1):386–402. https://doi.org/10.1172/JCI76738.
Jiang C, Xie C, Lv Y, Li J, Krausz KW, Shi J, et al. Intestine-selective farnesoid X receptor inhibition improves obesity-related metabolic dysfunction. Nat Commun. 2015;6:10166. https://doi.org/10.1038/ncomms10166.
Trabelsi MS, Daoudi M, Prawitt J, Ducastel S, Touche V, Sayin SI, et al. Farnesoid X receptor inhibits glucagon-like peptide-1 production by enteroendocrine L cells. Nat Commun. 2015;6:7629. https://doi.org/10.1038/ncomms8629.
Becker-Andre M, Andre E, DeLamarter JF. Identification of nuclear receptor mRNAs by RT-PCR amplification of conserved zinc-finger motif sequences. Biochem Biophys Res Commun. 1993;194(3):1371–9.
Chiang JY, Kimmel R, Weinberger C, Stroup D. Farnesoid X receptor responds to bile acids and represses cholesterol 7alpha-hydroxylase gene (CYP7A1) transcription. J Biol Chem. 2000;275(15):10918–24.
Galarneau L, Pare JF, Allard D, Hamel D, Levesque L, Tugwood JD, et al. The alpha1-fetoprotein locus is activated by a nuclear receptor of the Drosophila FTZ-F1 family. Mol Cell Biol. 1996;16(7):3853–65.
Goodwin B, Jones SA, Price RR, Watson MA, McKee DD, Moore LB, et al. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol Cell. 2000;6(3):517–26.
Lu TT, Makishima M, Repa JJ, Schoonjans K, Kerr TA, Auwerx J, et al. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol Cell. 2000;6(3):507–15.
Nitta M, Ku S, Brown C, Okamoto AY, Shan B. CPF: an orphan nuclear receptor that regulates liver-specific expression of the human cholesterol 7alpha-hydroxylase gene. Proc Natl Acad Sci U S A. 1999;96(12):6660–5.
Stroup D, Chiang JY. HNF4 and COUP-TFII interact to modulate transcription of the cholesterol 7alpha-hydroxylase gene (CYP7A1). J Lipid Res. 2000;41(1):1–11.
Zhang M, Chiang JY. Transcriptional regulation of the human sterol 12alpha-hydroxylase gene (CYP8B1): roles of heaptocyte nuclear factor 4alpha in mediating bile acid repression. J Biol Chem. 2001;276(45):41690–9. https://doi.org/10.1074/jbc.M105117200.
Lee YK, Moore DD. Dual mechanisms for repression of the monomeric orphan receptor liver receptor homologous protein-1 by the orphan small heterodimer partner. J Biol Chem. 2002;277(4):2463–7. https://doi.org/10.1074/jbc.M105161200.
Yang Y, Zhang M, Eggertsen G, Chiang JY. On the mechanism of bile acid inhibition of rat sterol 12alpha-hydroxylase gene (CYP8B1) transcription: roles of alpha-fetoprotein transcription factor and hepatocyte nuclear factor 4alpha. Biochim Biophys Acta. 2002;1583(1):63–73.
Holt JA, Luo G, Billin AN, Bisi J, McNeill YY, Kozarsky KF, et al. Definition of a novel growth factor-dependent signal cascade for the suppression of bile acid biosynthesis. Genes Dev. 2003;17(13):1581–91. https://doi.org/10.1101/gad.1083503.
Inagaki T, Choi M, Moschetta A, Peng L, Cummins CL, McDonald JG, et al. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2005;2(4):217–25. https://doi.org/10.1016/j.cmet.2005.09.001.
Kong B, Wang L, Chiang JY, Zhang Y, Klaassen CD, Guo GL. Mechanism of tissue-specific farnesoid X receptor in suppressing the expression of genes in bile-acid synthesis in mice. Hepatology. 2012;56(3):1034–43. https://doi.org/10.1002/hep.25740.
Song KH, Li T, Owsley E, Strom S, Chiang JY. Bile acids activate fibroblast growth factor 19 signaling in human hepatocytes to inhibit cholesterol 7alpha-hydroxylase gene expression. Hepatology. 2009;49(1):297–305. https://doi.org/10.1002/hep.22627.
Kerr TA, Saeki S, Schneider M, Schaefer K, Berdy S, Redder T, et al. Loss of nuclear receptor SHP impairs but does not eliminate negative feedback regulation of bile acid synthesis. Dev Cell. 2002;2(6):713–20.
Wang L, Lee YK, Bundman D, Han Y, Thevananther S, Kim CS, et al. Redundant pathways for negative feedback regulation of bile acid production. Dev Cell. 2002;2(6):721–31.
• Xu Y, Li F, Zalzala M, Xu J, Gonzalez FJ, Adorini L, et al. Farnesoid X receptor activation increases reverse cholesterol transport by modulating bile acid composition and cholesterol absorption in mice. Hepatology. 2016;64(4):1072–85. https://doi.org/10.1002/hep.28712 This study provides important in vivo tissue-specific information on FXR regulation of hepatic bile acid metabolism.
de Aguiar Vallim TQ, Tarling EJ, Ahn H, Hagey LR, Romanoski CE, Lee RG, et al. MAFG is a transcriptional repressor of bile acid synthesis and metabolism. Cell Metab. 2015;21(2):298–311. https://doi.org/10.1016/j.cmet.2015.01.007.
• Tarling EJ, Clifford BL, Cheng J, Morand P, Cheng A, Lester E, et al. RNA-binding protein ZFP36L1 maintains posttranscriptional regulation of bile acid metabolism. J Clin Invest. 2017;127(10):3741–54. https://doi.org/10.1172/JCI94029 This manuscript identifies an important post-transcriptional regulator of bile acid metabolism.
de Aguiar Vallim TQ, Tarling EJ, Kim T, Civelek M, Baldan A, Esau C, et al. MicroRNA-144 regulates hepatic ATP binding cassette transporter A1 and plasma high-density lipoprotein after activation of the nuclear receptor farnesoid X receptor. Circ Res. 2013;112(12):1602–12. https://doi.org/10.1161/CIRCRESAHA.112.300648.
Song KH, Li T, Owsley E, Chiang JY. A putative role of micro RNA in regulation of cholesterol 7alpha-hydroxylase expression in human hepatocytes. J Lipid Res. 2010;51(8):2223–33. https://doi.org/10.1194/jlr.M004531.
Li T, Francl JM, Boehme S, Chiang JY. Regulation of cholesterol and bile acid homeostasis by the cholesterol 7alpha-hydroxylase/steroid response element-binding protein 2/microRNA-33a axis in mice. Hepatology. 2013;58(3):1111–21. https://doi.org/10.1002/hep.26427.
Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, et al. Treatment of HCV infection by targeting microRNA. The N Engl J Med. 2013;368(18):1685–94. https://doi.org/10.1056/NEJMoa1209026.
Schmidt MF. miRNA targeting drugs: the next blockbusters? Methods Mol Biol. 2017;1517:3–22. https://doi.org/10.1007/978-1-4939-6563-2_1.
Baker DM, Wang SL, Bell DJ, Drevon CA, Davis RA. One or more labile proteins regulate the stability of chimeric mRNAs containing the 3′-untranslated region of cholesterol-7alpha -hydroxylase mRNA. J Biol Chem. 2000;275(26):19985–91. https://doi.org/10.1074/jbc.M002351200.
Noshiro M, Nishimoto M, Okuda K. Rat liver cholesterol 7 alpha-hydroxylase. Pretranslational regulation for circadian rhythm. J Biol Chem. 1990;265(17):10036–41.
Pandak WM, Stravitz RT, Lucas V, Heuman DM, Chiang JY. Hep G2 cells: a model for studies on regulation of human cholesterol 7alpha-hydroxylase at the molecular level. Am J Phys. 1996;270(3 Pt 1):G401–10. https://doi.org/10.1152/ajpgi.1996.270.3.G401.
Fiorucci S, Baldelli F. Farnesoid X receptor agonists in biliary tract disease. Curr Opin Gastroenterol. 2009;25(3):252–9. https://doi.org/10.1097/MOG.0b013e328324f87e.
Fiorucci S, Distrutti E, Ricci P, Giuliano V, Donini A, Baldelli F. Targeting FXR in cholestasis: hype or hope. Expert Opin Ther Targets. 2014;18(12):1449–59. https://doi.org/10.1517/14728222.2014.956087.
Fiorucci S, Mencarelli A, Cipriani S, Renga B, Palladino G, Santucci L, et al. Activation of the farnesoid-X receptor protects against gastrointestinal injury caused by non-steroidal anti-inflammatory drugs in mice. Br J Pharmacol. 2011;164(8):1929–38. https://doi.org/10.1111/j.1476-5381.2011.01481.x.
Fiorucci S, Mencarelli A, Distrutti E, Zampella A. Farnesoid X receptor: from medicinal chemistry to clinical applications. Future Med Chem. 2012;4(7):877–91. https://doi.org/10.4155/fmc.12.41.
Sepe V, Distrutti E, Fiorucci S, Zampella A. Farnesoid X receptor modulators (2011 - 2014): a patent review. Expert Opin Ther Pat. 2015;25(8):885–96. https://doi.org/10.1517/13543776.2015.1045413.
Erickson SK, Lear SR, Deane S, Dubrac S, Huling SL, Nguyen L, et al. Hypercholesterolemia and changes in lipid and bile acid metabolism in male and female cyp7A1-deficient mice. J Lipid Res. 2003;44(5):1001–9. https://doi.org/10.1194/jlr.M200489-JLR200.
Pullinger CR, Eng C, Salen G, Shefer S, Batta AK, Erickson SK, et al. Human cholesterol 7alpha-hydroxylase (CYP7A1) deficiency has a hypercholesterolemic phenotype. J Clin Invest. 2002;110(1):109–17. https://doi.org/10.1172/JCI15387.
Fiorucci S, Cipriani S, Baldelli F, Mencarelli A. Bile acid-activated receptors in the treatment of dyslipidemia and related disorders. Prog Lipid Res. 2010;49(2):171–85. https://doi.org/10.1016/j.plipres.2009.11.001.
Fiorucci S, Mencarelli A, Palladino G, Cipriani S. Bile-acid-activated receptors: targeting TGR5 and farnesoid-X-receptor in lipid and glucose disorders. Trends Pharmacol Sci. 2009;30(11):570–80. https://doi.org/10.1016/j.tips.2009.08.001.
Kern F Jr. Epidemiology and natural history of gallstones. Semin Liver Dis. 1983;3(2):87–96. https://doi.org/10.1055/s-2008-1040675.
Salen G, Nicolau G, Shefer S, Mosbach EH. Hepatic cholesterol metabolism in patients with gallstones. Gastroenterology. 1975;69(3):676–84.
Maruyama T, Miyamoto Y, Nakamura T, Tamai Y, Okada H, Sugiyama E, et al. Identification of membrane-type receptor for bile acids (M-BAR). Biochem Biophys Res Commun. 2002;298(5):714–9.
Kawamata Y, Fujii R, Hosoya M, Harada M, Yoshida H, Miwa M, et al. A G protein-coupled receptor responsive to bile acids. J Biol Chem. 2003;278(11):9435–40. https://doi.org/10.1074/jbc.M209706200.
Maruyama T, Tanaka K, Suzuki J, Miyoshi H, Harada N, Nakamura T, et al. Targeted disruption of G protein-coupled bile acid receptor 1 (Gpbar1/M-Bar) in mice. J Endocrinol. 2006;191(1):197–205. https://doi.org/10.1677/joe.1.06546.
Poole DP, Godfrey C, Cattaruzza F, Cottrell GS, Kirkland JG, Pelayo JC et al. Expression and function of the bile acid receptor GpBAR1 (TGR5) in the murine enteric nervous system. Neurogastroenterol Motil 2010;22(7):814–25, e227–8. doi:https://doi.org/10.1111/j.1365-2982.2010.01487.x, e228.
Keitel V, Reinehr R, Gatsios P, Rupprecht C, Gorg B, Selbach O, et al. The G-protein coupled bile salt receptor TGR5 is expressed in liver sinusoidal endothelial cells. Hepatology. 2007;45(3):695–704. https://doi.org/10.1002/hep.21458.
Keitel V, Ullmer C, Haussinger D. The membrane-bound bile acid receptor TGR5 (Gpbar-1) is localized in the primary cilium of cholangiocytes. Biol Chem. 2010;391(7):785–9. https://doi.org/10.1515/BC.2010.077.
Khurana S, Raufman JP, Pallone TL. Bile acids regulate cardiovascular function. Clin Transl Sci. 2011;4(3):210–8. https://doi.org/10.1111/j.1752-8062.2011.00272.x.
Keitel V, Donner M, Winandy S, Kubitz R, Haussinger D. Expression and function of the bile acid receptor TGR5 in Kupffer cells. Biochem Biophys Res Commun. 2008;372(1):78–84. https://doi.org/10.1016/j.bbrc.2008.04.171.
Yang JI, Yoon JH, Myung SJ, Gwak GY, Kim W, Chung GE, et al. Bile acid-induced TGR5-dependent c-Jun-N terminal kinase activation leads to enhanced caspase 8 activation in hepatocytes. Biochem Biophys Res Commun. 2007;361(1):156–61. https://doi.org/10.1016/j.bbrc.2007.07.001.
Giaretta PR, Suchodolski JS, Blick AK, Steiner JM, Lidbury JA, Rech RR. Distribution of bile acid receptor TGR5 in the gastrointestinal tract of dogs. Histol Histopathol. 2018:18025. https://doi.org/10.14670/HH-18-025.
Keitel V, Haussinger D. TGR5 in the biliary tree. Dig Dis. 2011;29(1):45–7. https://doi.org/10.1159/000324127.
Shaham O, Wei R, Wang TJ, Ricciardi C, Lewis GD, Vasan RS, et al. Metabolic profiling of the human response to a glucose challenge reveals distinct axes of insulin sensitivity. Mol Syst Biol. 2008;4:214. https://doi.org/10.1038/msb.2008.50.
Thomas C, Gioiello A, Noriega L, Strehle A, Oury J, Rizzo G, et al. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab. 2009;10(3):167–77. https://doi.org/10.1016/j.cmet.2009.08.001.
Brighton CA, Rievaj J, Kuhre RE, Glass LL, Schoonjans K, Holst JJ, et al. Bile acids trigger GLP-1 release predominantly by accessing basolaterally located G protein-coupled bile acid receptors. Endocrinology. 2015;156(11):3961–70. https://doi.org/10.1210/en.2015-1321.
Kuhre RE, Wewer Albrechtsen NJ, Larsen O, Jepsen SL, Balk-Moller E, Andersen DB, et al. Bile acids are important direct and indirect regulators of the secretion of appetite- and metabolism-regulating hormones from the gut and pancreas. Mol Metab. 2018;11:84–95. https://doi.org/10.1016/j.molmet.2018.03.007.
Carino A, Cipriani S, Marchiano S, Biagioli M, Scarpelli P, Zampella A, et al. Gpbar1 agonism promotes a Pgc-1alpha-dependent browning of white adipose tissue and energy expenditure and reverses diet-induced steatohepatitis in mice. Sci Rep. 2017;7(1):13689. https://doi.org/10.1038/s41598-017-13102-y.
Velazquez-Villegas LA, Perino A, Lemos V, Zietak M, Nomura M, Pols TWH, et al. TGR5 signalling promotes mitochondrial fission and beige remodelling of white adipose tissue. Nat Commun. 2018;9(1):245. https://doi.org/10.1038/s41467-017-02068-0.
Watanabe M, Houten SM, Mataki C, Christoffolete MA, Kim BW, Sato H, et al. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature. 2006;439(7075):484–9. https://doi.org/10.1038/nature04330.
Perino A, Pols TW, Nomura M, Stein S, Pellicciari R, Schoonjans K. TGR5 reduces macrophage migration through mTOR-induced C/EBPbeta differential translation. J Clin Invest. 2014;124(12):5424–36. https://doi.org/10.1172/JCI76289.
Pols TW, Nomura M, Harach T, Lo Sasso G, Oosterveer MH, Thomas C, et al. TGR5 activation inhibits atherosclerosis by reducing macrophage inflammation and lipid loading. Cell Metab. 2011;14(6):747–57. https://doi.org/10.1016/j.cmet.2011.11.006.
Alemi F, Kwon E, Poole DP, Lieu T, Lyo V, Cattaruzza F, et al. The TGR5 receptor mediates bile acid-induced itch and analgesia. J Clin Invest. 2013;123(4):1513–30. https://doi.org/10.1172/JCI64551.
Lieu T, Jayaweera G, Zhao P, Poole DP, Jensen D, Grace M, et al. The bile acid receptor TGR5 activates the TRPA1 channel to induce itch in mice. Gastroenterology. 2014;147(6):1417–28. https://doi.org/10.1053/j.gastro.2014.08.042.
Vassileva G, Golovko A, Markowitz L, Abbondanzo SJ, Zeng M, Yang S, et al. Targeted deletion of Gpbar1 protects mice from cholesterol gallstone formation. Biochem J. 2006;398(3):423–30. https://doi.org/10.1042/BJ20060537.
Briere DA, Ruan X, Cheng CC, Siesky AM, Fitch TE, Dominguez C, et al. Novel small molecule agonist of TGR5 possesses anti-diabetic effects but causes gallbladder filling in mice. PLoS One. 2015;10(8):e0136873. https://doi.org/10.1371/journal.pone.0136873.
Li T, Holmstrom SR, Kir S, Umetani M, Schmidt DR, Kliewer SA, et al. The G protein-coupled bile acid receptor, TGR5, stimulates gallbladder filling. Mol Endocrinol. 2011;25(6):1066–71. https://doi.org/10.1210/me.2010-0460.
Donepudi AC, Boehme S, Li F, Chiang JY. G-protein-coupled bile acid receptor plays a key role in bile acid metabolism and fasting-induced hepatic steatosis in mice. Hepatology. 2017;65(3):813–27. https://doi.org/10.1002/hep.28707.
Adams TD, Davidson LE, Litwin SE, Kolotkin RL, LaMonte MJ, Pendleton RC, et al. Health benefits of gastric bypass surgery after 6 years. JAMA. 2012;308(11):1122–31. https://doi.org/10.1001/2012.jama.11164.
Schauer PR, Bhatt DL, Kirwan JP, Wolski K, Brethauer SA, Navaneethan SD, et al. Bariatric surgery versus intensive medical therapy for diabetes--3-year outcomes. The N Engl J Med. 2014;370(21):2002–13. https://doi.org/10.1056/NEJMoa1401329.
Ding L, Sousa KM, Jin L, Dong B, Kim BW, Ramirez R, et al. Vertical sleeve gastrectomy activates GPBAR-1/TGR5 to sustain weight loss, improve fatty liver, and remit insulin resistance in mice. Hepatology. 2016;64(3):760–73. https://doi.org/10.1002/hep.28689.
Ryan KK, Tremaroli V, Clemmensen C, Kovatcheva-Datchary P, Myronovych A, Karns R, et al. FXR is a molecular target for the effects of vertical sleeve gastrectomy. Nature. 2014;509(7499):183–8. https://doi.org/10.1038/nature13135.
Bhutta HY, Rajpal N, White W, Freudenberg JM, Liu Y, Way J, et al. Effect of Roux-en-Y gastric bypass surgery on bile acid metabolism in normal and obese diabetic rats. PLoS One. 2015;10(3):e0122273. https://doi.org/10.1371/journal.pone.0122273.
Gerhard GS, Styer AM, Wood GC, Roesch SL, Petrick AT, Gabrielsen J, et al. A role for fibroblast growth factor 19 and bile acids in diabetes remission after Roux-en-Y gastric bypass. Diabetes Care. 2013;36(7):1859–64. https://doi.org/10.2337/dc12-2255.
Patti ME, Houten SM, Bianco AC, Bernier R, Larsen PR, Holst JJ, et al. Serum bile acids are higher in humans with prior gastric bypass: potential contribution to improved glucose and lipid metabolism. Obesity (Silver Spring). 2009;17(9):1671–7. https://doi.org/10.1038/oby.2009.102.
Pournaras DJ, Glicksman C, Vincent RP, Kuganolipava S, Alaghband-Zadeh J, Mahon D, et al. The role of bile after Roux-en-Y gastric bypass in promoting weight loss and improving glycaemic control. Endocrinology. 2012;153(8):3613–9. https://doi.org/10.1210/en.2011-2145.
Simonen M, Dali-Youcef N, Kaminska D, Venesmaa S, Kakela P, Paakkonen M, et al. Conjugated bile acids associate with altered rates of glucose and lipid oxidation after Roux-en-Y gastric bypass. Obes Surg. 2012;22(9):1473–80. https://doi.org/10.1007/s11695-012-0673-5.
Steinert RE, Peterli R, Keller S, Meyer-Gerspach AC, Drewe J, Peters T, et al. Bile acids and gut peptide secretion after bariatric surgery: a 1-year prospective randomized pilot trial. Obesity (Silver Spring). 2013;21(12):E660–8. https://doi.org/10.1002/oby.20522.
Kohli R, Bradley D, Setchell KD, Eagon JC, Abumrad N, Klein S. Weight loss induced by Roux-en-Y gastric bypass but not laparoscopic adjustable gastric banding increases circulating bile acids. J Clin Endocrinol Metab. 2013;98(4):E708–12. https://doi.org/10.1210/jc.2012-3736.
Kohli R, Setchell KD, Kirby M, Myronovych A, Ryan KK, Ibrahim SH, et al. A surgical model in male obese rats uncovers protective effects of bile acids post-bariatric surgery. Endocrinology. 2013;154(7):2341–51. https://doi.org/10.1210/en.2012-2069.
Flynn CR, Albaugh VL, Cai S, Cheung-Flynn J, Williams PE, Brucker RM, et al. Bile diversion to the distal small intestine has comparable metabolic benefits to bariatric surgery. Nat Commun. 2015;6:7715. https://doi.org/10.1038/ncomms8715 https://www.nature.com/articles/ncomms8715#supplementary-information.
Myronovych A, Salazar-Gonzalez RM, Ryan KK, Miles L, Zhang W, Jha P, et al. The role of small heterodimer partner in nonalcoholic fatty liver disease improvement after sleeve gastrectomy in mice. Obesity (Silver Spring). 2014;22(11):2301–11. https://doi.org/10.1002/oby.20890.
Habegger KM, Al-Massadi O, Heppner KM, Myronovych A, Holland J, Berger J, et al. Duodenal nutrient exclusion improves metabolic syndrome and stimulates villus hyperplasia. Gut. 2014;63(8):1238–46. https://doi.org/10.1136/gutjnl-2013-304583.
Kohli R, Kirby M, Setchell KD, Jha P, Klustaitis K, Woollett LA, et al. Intestinal adaptation after ileal interposition surgery increases bile acid recycling and protects against obesity-related comorbidities. Am J Physiol Gastrointest Liver Physiol. 2010;299(3):G652–60. https://doi.org/10.1152/ajpgi.00221.2010.
Hao Z, Leigh Townsend R, Mumphrey MB, Gettys TW, Yu S, Munzberg H, et al. Roux-en-Y gastric bypass surgery-induced weight loss and metabolic improvements are similar in TGR5-deficient and wildtype mice. Obes Surg. 2018. https://doi.org/10.1007/s11695-018-3297-6.
Zhai H, Li Z, Peng M, Huang Z, Qin T, Chen L, et al. Takeda G protein-coupled receptor 5-mechanistic target of rapamycin complex 1 signaling contributes to the increment of glucagon-like peptide-1 production after Roux-en-Y gastric bypass. EBioMedicine. 2018;32:201–14. https://doi.org/10.1016/j.ebiom.2018.05.026.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Karolina E. Zaborska and Bethany P. Cummings declare no conflicts of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
This article is part of the Topical Collection on Pathogenesis of Type 2 Diabetes and Insulin Resistance
Rights and permissions
About this article
Cite this article
Zaborska, K.E., Cummings, B.P. Rethinking Bile Acid Metabolism and Signaling for Type 2 Diabetes Treatment. Curr Diab Rep 18, 109 (2018). https://doi.org/10.1007/s11892-018-1092-3
Published:
DOI: https://doi.org/10.1007/s11892-018-1092-3