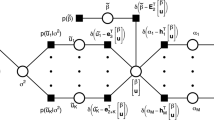
Abstract
This paper develops a methodology for robust Bayesian inference through the use of disparities. Metrics such as Hellinger distance and negative exponential disparity have a long history in robust estimation in frequentist inference. We demonstrate that an equivalent robustification may be made in Bayesian inference by substituting an appropriately scaled disparity for the log likelihood to which standard Monte Carlo Markov Chain methods may be applied. A particularly appealing property of minimum-disparity methods is that while they yield robustness with a breakdown point of 1/2, the resulting parameter estimates are also efficient when the posited probabilistic model is correct. We demonstrate that a similar property holds for disparity-based Bayesian inference. We further show that in the Bayesian setting, it is also possible to extend these methods to robustify regression models, random effects distributions and other hierarchical models. These models require integrating out a random effect; this is achieved via MCMC but would otherwise be numerically challenging. The methods are demonstrated on real-world data.



Similar content being viewed by others
References
Albert J (2008) LearnBayes: functions for learning Bayesian inference. R package version 2
Albert J (2009) Bayesian computation with R. Springer, New York
Andrade JAA, O’Hagan A (2006) Bayesian robustness modeling using regularly varying distributions. Bayesian Anal 1(1):169–188
Basu A, Sarkar S, Vidyashankar AN (1997) Minimum negative exponential disparity estimation in parametric models. J Stat Plan Inference 58:349–370
Basu A, Shioya H, Park C (2011) Statistical inference, monographs on statistics and applied probability, vol 120. CRC Press, Boca Raton (the minimum distance approach)
Beran R (1977) Minimum Hellinger distance estimates for parametric models. Ann Stat 5:445–463
Berger JO (1994) An overview of robust Bayesian analysis. TEST 3:5–124
Cheng AL, Vidyashankar AN (2006) Minimum Hellinger distance estimation for randomized play the winner design. J Stat Plan Inference 136:1875–1910
Choy STB, Smith AFM (1997) On robust analysis of a normal location parameter. J Royal Stat Soc B 59:463–474
Dawid AP (1973) Posterior expectations for large observations. Biometrika 60:664–667
Desgagnè A, Angers JF (2007) Confilicting information and location parameter inference. Metron 65:67–97
Devroye L, Györfi G (1985) Nonparametric density estimation: the L1 view. Wiley, New York
Dey DK, Birmiwal LR (1994) Robust Bayesian analysis using divergence measures. Stat Prob Lett 20: 287–294
Dunson DB, Taylor JA (2005) Approximate bayesian inference for quantiles. J Nonparametr Stat 17(3): 385–400
Engel J, Herrmann E, Gasser T (1994) An iterative bandwidth selector for kernel estimation of densities and their derivatives. J Nonparametr Stat 4:2134
Ghosh JK, Delampady M, Samanta T (2006) An introduction to Bayesian analysis. Springer, New York
Hampel FR (1974) The influence curve and its role in robust estimation. J Am Stat Assoc 69:383–393
Hampel FR, Ronchetti EM, Rousseeuw PJ, Stahel WA (1986) Robust statistics. Wiley Series in probability and mathematical statistics: probability and mathematical statistics. Wiley, New York (the approach based on influence functions)
Hansen BE (2004) Nonparametric conditional density estimation. http://www.ssc.wisc.edu/~bhansen/papers/ncde (Unpublished Manuscript)
Hoff PD (2007) Extending the rank likelihood for semiparametric copula estimation. Ann Appl Stat 1(1): 265–283
Hooker G (2013) Consistency, efficiency and robustness of conditional disparity methods. arXiv:1307.3730
Huber P (1981) Robust statistics. Wiley, New York
Jiang W, Tanner MA (2008) Gibbs posterior for variable selection in high-dimensional classification and data mining. Ann Stat 26(5):2207–2231
Jureckova J, Sen PK (1996) Robust statistical procedures. Wiley Series in probability and statistics: applied probability and statistics. Wiley, New York (asymptotics and interrelations, A Wiley-Interscience Publication)
Li Q, Racine JS (2007) Nonparametric econometrics. Princeton University Press, Princeton
Lindsay BG (1994) Efficiency versus robustness: the case for minimum Hellinger distance and related methods. Ann Stat 22:1081–1114
Maronna RA, Martin RD, Yohai VJ (2006) Robust statistics. Wiley Series in probability and statistics. Theory and methods. Wiley , Chichester
Nielsen M, Vidyashankar A, Hanlon B, Diao G, Petersen S, Kaplan R (2013) Hierarchical model for evaluating pyrantel efficacy against strongyle parasites in horses. Vet Parasitol 197(3):614–622
O’Hagan A (1979) On outlier rejection phenomena in bayes inference. J Royal Stat Soc B 41:358–367
O’Hagan A (1990) Outliers and credence for location parameter inference. J Am Stat Assoc 85:172–176
Park C, Basu A (2004) Minimum disparity estimation: asymptotic normality and breakdown point results. Bull Inf Cybernet 36:19–34
Peng F, Dey DK (1995) Bayesian analysis of outlier problems using divergence measures. Can J Stat 23:199–213
Sheather SJ, Jones MC (1991) A reliable data-based bandwidth selection method for kernel density estimation. J Royal Stat Soc Ser B 53:683690
Silverman BW (1982) Density estimation. Chapman and Hall, Boca Raton
Simpson DG (1987) Minimum Hellinger distance estimation for the analysis of count data. J Am Stat Assoc 82:802–807
Simpson DG (1989) Hellinger deviance test: efficiency, breakdown points and examples. J Am Stat Assoc 84:107–113
Sollich P (2002) Bayesian methods for support vector machines: evidence and predicive class probabilities. Mach Learn 46:21–52
Stigler SM (1973) The asymptotic distribution of the trimmed mean. Ann Stat 1:427–477
Szpiro AA, Rice KM, Lumley T (2010) Model-robust regression and a Bayesian “sandwich” estimator. Ann Appl Stat 4:2099–2113
Tamura RN, Boos DD (1986) Minimum Hellinger distances estimation for multivariate location and and covariance. J Am Stat Assoc 81:223–229
Wand M, Ripley B (2009) KernSmooth: functions for kernel smoothing. R package version 2.23-3
Wu Y, Hooker G (2013) Bayesian model robustness via disparities. arXiv:1112.4213
Zhan X, Hettmansperger TP (2007) Bayesian \(R\)-estimates in two-sample location models. Comput Statist Data Anal 51(10):5077–5089
Acknowledgments
Giles Hooker’s research was supported by National Science Foundation grant DEB-0813734 and the Cornell University Agricultural Experiment Station federal formula funds Project No. 150446. Anand N. Vidyashankar’s research was supported in part by a grant from National Science Foundation, DMS-000-03-07057.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Appendix
Appendix
Appendices A–F are available as Online Resource 1.
Rights and permissions
About this article
Cite this article
Hooker, G., Vidyashankar, A.N. Bayesian model robustness via disparities. TEST 23, 556–584 (2014). https://doi.org/10.1007/s11749-014-0360-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11749-014-0360-z
Keywords
- Deviance test
- Kernel density
- Hellinger distance
- Negative exponential disparity
- MCMC
- Bayesian inference
- Posterior
- Outliers