Abstract
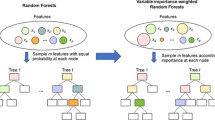
Random forests are a commonly used tool for classification and for ranking candidate predictors based on the so-called variable importance measures. These measures attribute scores to the variables reflecting their importance. A drawback of variable importance measures is that there is no natural cutoff that can be used to discriminate between important and non-important variables. Several approaches, for example approaches based on hypothesis testing, were developed for addressing this problem. The existing testing approaches require the repeated computation of random forests. While for low-dimensional settings those approaches might be computationally tractable, for high-dimensional settings typically including thousands of candidate predictors, computing time is enormous. In this article a computationally fast heuristic variable importance test is proposed that is appropriate for high-dimensional data where many variables do not carry any information. The testing approach is based on a modified version of the permutation variable importance, which is inspired by cross-validation procedures. The new approach is tested and compared to the approach of Altmann and colleagues using simulation studies, which are based on real data from high-dimensional binary classification settings. The new approach controls the type I error and has at least comparable power at a substantially smaller computation time in the studies. Thus, it might be used as a computationally fast alternative to existing procedures for high-dimensional data settings where many variables do not carry any information. The new approach is implemented in the R package vita.








Similar content being viewed by others
References
Alon U, Barkai N, Notterman DA, Gish K, Ybarra S, Mack D, Levine AJ (1999) Broad patterns of gene expression revealed by clustering analysis of tumor and normal colon tissues probed by oligonucleotide arrays. Proc Natl Acad Sci 96:6745–6750
Altmann A, Toloşi L, Sander O, Lengauer T (2010) Permutation importance: a corrected feature importance measure. Bioinformatics 26:1340–1347
Boulesteix A-L (2015) Ten simple rules for reducing overoptimistic reporting in methodological computational research. PLoS Comput Biol 4:e1004191
Boulesteix AL, Bender A, Bermejo JL, Strobl C (2012) Random forest Gini importance favours SNPs with large minor allele frequency: assessment, sources and recommendations. Brief Bioinform 13:292–304
Breiman L (2001) Random forests. Mach Learn 45:5–32
Breiman L, C. A (2008) Random forests. http://www.stat.berkeley.edu/users/breiman/RandomForests/cc_home.htm
Dettling M, Bühlmann P (2003) Boosting for tumor classification with gene expression data. Bioinformatics 19:1061–1069
Díaz-Uriarte R, De Andres SA (2006) Gene selection and classification of microarray data using random forest. BMC Bioinform 7:3
Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA et al (1999) Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 286:531–537
Gregorutti B, Michel B, Saint-Pierre P (2013) Correlation and variable importance in random forests. arXiv preprint arXiv:1310.5726
Hapfelmeier A, Ulm K (2013) A new variable selection approach using random forests. Comput Stat Data Anal 60:50–69
Hothorn T, Hornik K, Zeileis A (2006) Unbiased recursive partitioning: a conditional inference framework. J Comput Graph Stat 15:651–674
Huynh-Thu VA, Saeys Y, Wehenkel L, Geurts P (2012) Statistical interpretation of machine learning-based feature importance scores for biomarker discovery. Bioinformatics 28:1766–1774
Ishwaran H (2007) Variable importance in binary regression trees and forests. Electron J Stat 1:519–537
Ishwaran H, Kogalur UB, Blackstone EH, Lauer MS (2008) Random survival forests. Ann Appl Stat 2:841–860
Janitza S, Strobl C, Boulesteix AL (2013) An AUC-based permutation variable importance measure for random forests. BMC Bioinform 14:119
Janitza S, Tutz G, Boulesteix A-L (2016) Random forest for ordinal responses: prediction and variable selection. Comput Stat Data Anal 96:57–73
Kim H, Loh W-Y (2001) Classification trees with unbiased multiway splits. J Am Stat Assoc 96:589–604
Liaw A, Wiener M (2002) Classification and regression by randomForest. R News 2:18–22
Louppe G, Wehenkel L, Sutera A, Geurts P (2013) Understanding variable importances in forests of randomized trees. In: Burges CJC, Bottou L, Welling M, Ghahramani Z, Weinberger KQ (eds) Advances in neural information processing systems, pp 431–439
Molinaro AM, Carriero N, Bjornson R, Hartge P, Rothman N, Chatterjee N (2011) Power of data mining methods to detect genetic associations and interactions. Hum Hered 72:85–97
Nicodemus K (2011) Letter to the editor: on the stability and ranking of predictors from random forest variable importance measures. Brief Bioinform 12:369–373
Nicodemus K, Malley J (2009) Predictor correlation impacts machine learning algorithms: implications for genomic studies. Bioinformatics 25:1884–1890
Pepe M (2004) The statistical evaluation of medical tests for classification and prediction. Oxford University Press, USA
Phipson B, Smyth G (2010) Permutation P-values should never be zero: calculating exact P-values when permutations are randomly drawn. Stat Appl Genet Mol Biol 9:1544–6115
Polak P, Karlić R, Koren A, Thurman R, Sandstrom R, Lawrence MS, Reynolds A, Rynes E, Vlahoviček K, Stamatoyannopoulos JA et al (2015) Cell-of-origin chromatin organization shapes the mutational landscape of cancer. Nature 518:360–364
Pomeroy SL, Tamayo P, Gaasenbeek M, Sturla LM, Angelo M, McLaughlin ME, Kim JY, Goumnerova LC, Black PM, Lau C et al (2002) Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature 415:436–442
Prosperi MC, Marinho S, Simpson A, Custovic A, Buchan IE (2014) Predicting phenotypes of asthma and eczema with machine learning. BMC Med Genomics 7:S7
Reif DM, Motsinger-Reif AA, McKinney BA, Rock MT, Crowe J, Moore JH (2009) Integrated analysis of genetic and proteomic data identifies biomarkers associated with adverse events following smallpox vaccination. Genes Immun 10:112–119
Schwarz DF, König IR, Ziegler A (2010) On safari to random jungle: a fast implementation of random forests for high-dimensional data. Bioinformatics 26:1752–1758
Singh D, Febbo PG, Ross K, Jackson DG, Manola J, Ladd C, Tamayo P, Renshaw AA, D’Amico AV, Richie JP et al (2002) Gene expression correlates of clinical prostate cancer behavior. Cancer Cell 1:203–209
Strobl C, Boulesteix A-L, Kneib T, Augustin T, Zeileis A (2008) Conditional variable importance for random forests. BMC Bioinform 9:307
Strobl C, Boulesteix AL, Zeileis A, Hothorn T (2007) Bias in random forest variable importance measures: Illustrations, sources and a solution. BMC Bioinform 8:25
Strobl C, Malley J, Tutz G (2009) An introduction to recursive partitioning: rationale, application, and characteristics of classification and regression trees, bagging, and random forests. Psychol Methods 14:323–348
Strobl C, Zeileis A (2008) Danger: high power!—exploring the statistical properties of a test for random forest variable importance. In: Brito P (ed) Proceedings of the 18th international conference on computational statistics. Porto, Portugal (CD-ROM), Springer, Heidelberg, pp 59–66
Szymczak S, Holzinger E, Dasgupta A, Malley JD, Molloy AN, Mills JL, Brody LC, Stambolian D, Bailey-Wilson JE (2016) r2VIM: a new variable selection method for random forests in genome-wide association studies. BioData Min 9:7
Tan AC, Gilbert D (2003) Ensemble machine learning on gene expression data for cancer classification. Appl Bioinform 2:S75–S83
Tang R, Sinnwell JP, Li J, Rider DN, de Andrade M, Biernacka JM (2009) Identification of genes and haplotypes that predict rheumatoid arthritis using random forests. BMC Proc 3:S68
van’t Veer LJ, Dai H, Van De Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT et al (2002) Gene expression profiling predicts clinical outcome of breast cancer. Nature 415:530–536
Wang H, Yang F, Luo Z (2016) An experimental study of the intrinsic stability of random forest variable importance measures. BMC Bioinform 17:60
Wang-Sattler R, Yu Z, Herder C, Messias AC, Floegel A, He Y, Heim K, Campillos M, Holzapfel C, Thorand B et al (2012) Novel biomarkers for pre-diabetes identified by metabolomics. Mol Syst Biol 8:615. doi:10.1038/msb.2012.43
Wright MN, Ziegler A (2016) ranger: a fast implementation of random forests for high dimensional data in C++ and R. J Stat Softw (in press)
Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP et al (2012) Human gut microbiome viewed across age and geography. Nature 486:222–227
Zhu R, Zeng D, Kosorok MR (2015) Reinforcement learning trees. JASA 110:1770–1784
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Janitza, S., Celik, E. & Boulesteix, AL. A computationally fast variable importance test for random forests for high-dimensional data. Adv Data Anal Classif 12, 885–915 (2018). https://doi.org/10.1007/s11634-016-0276-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11634-016-0276-4
Keywords
- Gene selection
- Feature selection
- Random forests
- Variable importance
- Variable selection
- Variable importance test