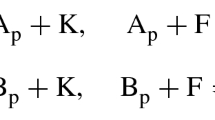Abstract
This work addresses whether a reaction network, taken with mass-action kinetics, is multistationary, that is, admits more than one positive steady state in some stoichiometric compatibility class. We build on previous work on the effect that removing or adding intermediates has on multistationarity, and also on methods to detect multistationarity for networks with a binomial steady-state ideal. In particular, we provide a new determinant criterion to decide whether a network is multistationary, which applies when the network obtained by removing intermediates has a binomial steady-state ideal. We apply this method to easily characterize which subsets of complexes are responsible for multistationarity; this is what we call the multistationarity structure of the network. We use our approach to compute the multistationarity structure of the n-site sequential distributive phosphorylation cycle for arbitrary n.
Similar content being viewed by others
References
Basu S, Pollack R, Coste-Roy MF (2007) Algorithms in real algebraic geometry, vol 10. Springer, Berlin
Bihan F, Dickenstein A, Giaroli M (2019) Lower bounds for positive roots and regions of multistationarity in chemical reaction networks. arXiv:1807.05157
Bradford R, Davenport JH, England M, Errami H, Gerdt V, Grigoriev D, Hoyt C, Košta M, Radulescu O, Sturm T, Weber A (2017) A case study on the parametric occurrence of multiple steady states. In: Proceedings of the international symposium on symbolic and algebraic computation, ISSAC, pp 45–52. Association for Computing Machinery
Dickenstein A, Pérez Millán M, Shiu A, Tang X (2019) Multistationarity in structured reaction networks. Bull Math Biol 81:1527–1581
Eisenbud D, Sturmfels B (1996) Binomial ideals. Duke Math J 84(1):1–45
England M, Bradford R, Davenport JH (2015) Improving the use of equational constraints in cylindrical algebraic decomposition. In: Proceedings of the international symposium on symbolic and algebraic computation, ISSAC, pp 165–172. Association for Computing Machinery
Feinberg M (1980) Lectures on chemical reaction networks. http://www.crnt.osu.edu/LecturesOnReactionNetworks
Feinberg M (1995) The existence and uniqueness of steady states for a class of chemical reaction networks. Arch Ration Mech Anal 132(4):311–370
Feliu E, Wiuf C (2012) Enzyme-sharing as a cause of multi-stationarity in signalling systems. J R Soc Interface 9(71):1224–1232
Feliu E, Wiuf C (2013) Simplifying biochemical models with intermediate species. J R Soc Interface 10(87):20130484
Gerhard J, Jeffrey D, Moroz G (2010) A package for solving parametric polynomial systems. ACM Commun Comput Algebra 43(3/4):61–72
Gunawardena J (2003) Chemical reaction network theory for in-silico biologists. http://vcp.med.harvard.edu/papers/crnt.pdf
Lazard D, Rouillier F (2007) Solving parametric polynomial systems. J Symb Comput 42(6):636–667
Müller S, Feliu E, Regensburger G, Conradi C, Shiu A, Dickenstein A (2016) Sign conditions for injectivity of generalized polynomial maps with applications to chemical reaction networks and real algebraic geometry. Found Comput Math 16(1):69–97
Pérez Millán M, Dickenstein A (2018) The structure of MESSI biological systems. SIAM J Appl Dyn Syst 17(2):1650–1682
Pérez Millán M, Dickenstein A, Shiu A, Conradi C (2012) Chemical reaction systems with toric steady states. Bull Math Biol 74(5):1027–1065
Sadeghimanesh AH, Feliu E (2019) Gröbner bases of reaction networks with intermediate species. Adv Appl Math 107(2):74–101
Sáez M, Wiuf C, Feliu E (2017) Graphical reduction of reaction networks by linear elimination of species. J Math Biol 74(1):195–237
Schilling CH, Letscher D, Palsson BØ (2000) Theory for the systemic definition of metabolic pathways and their use in interpreting metabolic function from a pathway-oriented perspective. J Theor Biol 203(3):229–248
Wang L, Sontag ED (2008) On the number of steady states in a multiple futile cycle. J Math Biol 57(1):29–52
Acknowledgements
This work has been supported by the Independent Research Fund of Denmark. We thank Alicia Dickenstein, Martin Helmer and Angélica Torres for comments on a preliminary version of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sadeghimanesh, A., Feliu, E. The Multistationarity Structure of Networks with Intermediates and a Binomial Core Network. Bull Math Biol 81, 2428–2462 (2019). https://doi.org/10.1007/s11538-019-00612-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11538-019-00612-1