Abstract
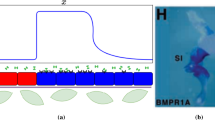
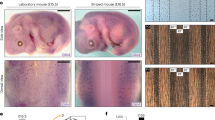
Here, we present a theoretical investigation with potential insights on developmental mechanisms. Three biological factors, consisting of two diffusing factors and a cell-autonomous immobile transcription factor are combined with different feedback mechanisms. This results in four different situations or fur patterns. Two of them reproduce classical Turing patterns: (1) regularly spaced spots, (2) labyrinth patterns or straight lines with an initial slope in the activation of the transcription factor. The third situation does not lead to patterns, but results in different homogeneous color tones. Finally, the fourth one sheds new light on the possible mechanisms leading to the formation of piebald patterns exemplified by the random patterns on the fur of some cows’ strains and Dalmatian dogs. Piebaldism is usually manifested as white areas of fur, hair, or skin due to the absence of pigment-producing cells in those regions. The distribution of the white and colored zones does not reflect the classical Turing patterns. We demonstrate that these piebald patterns are of transient nature, developing from random initial conditions and relying on a system’s bistability. We show numerically that the presence of a cell-autonomous factor not only expands the range of reaction diffusion parameters in which a pattern may arise, but also extends the pattern-forming abilities of the reaction–diffusion equations.






Similar content being viewed by others
References
Abdel-Malek ZA, Swope VB (2011) Epidermal melanocytes: regulation of their survival, proliferation, and function in human skin. In: Bosserhoff A (ed) Melanoma development: molecular biology, genetics and clinical application. Springer, Vienna, pp 7–33
Adcock IM, Caramori G (2009) Chapter 31—transcription factors A2—Barnes, Peter J. In: Drazen JM, Rennard SI, Thomson NC (eds) Asthma and COPD, 2nd edn. Academic Press, Oxford, pp 373–380
Allen WL, Cuthill IC, Scott-Samuel NE, Baddeley R (2011) Why the leopard got its spots: relating pattern development to ecology in felids. Proc R Soc B Biol Sci 278:1373–1380
Anma A, Sakamoto K, Yoneda T (2012) Unstable subsystems cause Turing instability. Kodai Math J 35:215–247
Attie T, Till M, Pelet A, Amiel J, Edery P, Boutrand L, Munnich A, Lyonnet S (1995) Mutation of the endothelin-receptor B gene in Waardenburg-Hirschsprung disease. Hum Mol Genet 4:2407–2409
Baker JC, Beddington RSP, Harland RM (1999) Wnt signaling in Xenopus embryos inhibits Bmp4 expression and activates neural development. Genes Dev 13:3149–3159
Basu S, Gerchman Y, Collins CH, Arnold FH, Weiss R (2005) A synthetic multicellular system for programmed pattern formation. Nature 434:1130–1134
Cai L, Dalal CK, Elowitz MB (2008) Frequency-modulated nuclear localization bursts coordinate gene regulation. Nature 455:485–490
Cao Y, Ryser MD, Payne S, Li B, Rao CV, You L (2016) Collective space-sensing coordinates pattern scaling in engineered bacteria. Cell 165:620–630
Cichorek M, Wachulska M, Stasiewicz A, Tymińska A (2013) Skin melanocytes: biology and development. Adv Dermatol Allergol 30:30–41
Danino T, Mondragon-Palomino O, Tsimring L, Hasty J (2010) A synchronized quorum of genetic clocks. Nature 463:326–330
Egri Á, Blahó M, Kriska G, Farkas R, Gyurkovszky M, Åkesson S, Horváth G (2012) Polarotactic tabanids find striped patterns with brightness and/or polarization modulation least attractive: an advantage of zebra stripes. J Exp Biol 215:736–745
Filtz TM, Vogel WK, Leid M (2014) Regulation of transcription factor activity by interconnected, post-translational modifications. Trends Pharmacol Sci 35:76–85
Frances L, Betlloch I, Leiva-Salinas M, Silvestre JF (2015) Spontaneous repigmentation in an infant with piebaldism. Int J Dermatol 54:e244–e246
Griffiths C, Barker J, Bleiker T, Chalmers R, Creamer D (2016) Rook’s textbook of dermatology, vol 4. Wiley, Hoboken
Hirobe T (2011) How are proliferation and differentiation of melanocytes regulated? Pigment Cell Melanoma Res 24:462–478
Hiscock TW, Megason SG (2015) Orientation of turing-like patterns by morphogen gradients and tissue anisotropies. Cell Syst 1:408–416
How MJ, Zanker JM (2014) Motion camouflage induced by zebra stripes. Zoology 117:163–170
Huang A, Glick SA (2016) Piebaldism in history-”The Zebra People”. JAMA Dermatol 152:1261
Kaelin CB, Xu X, Hong LZ, David VA, McGowan KA, Schmidt-Kuntzel A, Roelke ME, Pino J, Pontius J, Cooper GM et al (2012) Specifying and sustaining pigmentation patterns in domestic and wild cats. Science 337:1536–1541
Klika V, Baker RE, Headon D, Gaffney EA (2012) The influence of receptor-mediated interactions on reaction-diffusion mechanisms of cellular self-organisation. Bull Math Biol 74:935–957
Kondo S, Asai R (1995) A reaction–diffusion wave on the skin of the marine angelfish Pomacanthus. Nature 376:765
Korvasova K, Gaffney EA, Maini PK, Ferreira MA, Klika V (2015) Investigating the Turing conditions for diffusion-driven instability in the presence of a binding immobile substrate. J Theor Biol 367:286–295
Li A, Ma Y, Yu X, Mort RL, Lindsay CR, Stevenson D, Strathdee D, Insall RH, Chernoff J, Snapper SB et al (2011) Rac1 drives melanoblast organization during mouse development by orchestrating pseudopod-driven motility and cell-cycle progression. Dev Cell 21:722–734
Lin JY, Fisher DE (2007) Melanocyte biology and skin pigmentation. Nature 445:843–850
Liu C, Fu X, Liu L, Ren X, Chau CKL, Li S, Xiang L, Zeng H, Chen G, Tang L-H et al (2011) Sequential establishment of stripe patterns in an expanding cell population. Science 334:238
Mallarino R, Henegar C, Mirasierra M, Manceau M, Schradin C, Vallejo M, Beronja S, Barsh GS, Hoekstra HE (2016) Developmental mechanisms of stripe patterns in rodents. Nature 539:518–523
Marciniak-Czochra A (2003) Receptor-based models with diffusion-driven instability for pattern formation in hydra. J Biol Syst 11:293–324
Marcon L, Diego X, Sharpe J, Müller P (2016) High-throughput mathematical analysis identifies Turing networks for patterning with equally diffusing signals. eLife 5:e14022
McMahon JA, Takada S, Zimmerman LB, Fan C-M, Harland RM, McMahon AP (1998) Noggin-mediated antagonism of BMP signaling is required for growth and patterning of the neural tube and somite. Genes Dev 12:1438–1452
Mills MG, Patterson LB (2009) Not just black and white: pigment pattern development and evolution in vertebrates. Semin Cell Dev Biol 20:72–81
Miura, T. (2007). Modulation of activator diffusion by extracellular matrix in Turing system (Workshops on “Pattern Formation Problems in Dissipative Systems” and “Mathematical Modeling and Analysis for Nonlinear Phenomena”). 数理解析研究所講究録別冊 = RIMS Kokyuroku Bessatsu B3:165–176
Miyazawa S, Okamoto M, Kondo S (2010) Blending of animal colour patterns by hybridization. Nat Commun 1:66
Mort RL, Ross RJ, Hainey KJ, Harrison OJ, Keighren MA, Landini G, Baker RE, Painter KJ, Jackson IJ, Yates CA (2016) Reconciling diverse mammalian pigmentation patterns with a fundamental mathematical model. Nat Commun 7:10288
Murray JD (1989) Biological waves: multi-species reaction diffusion models. In: Mathematical biology, Springer, Berlin, pp 311–359
Nakayama K, Satoh T, Igari A, Kageyama R, Nishida E (2008) FGF induces oscillations of Hes1 expression and Ras/ERK activation. Curr Biol CB 18:R332–R334
Nelson DE, Ihekwaba AE, Elliott M, Johnson JR, Gibney CA, Foreman BE, Nelson G, See V, Horton CA, Spiller DG et al (2004) Oscillations in NF-kappaB signaling control the dynamics of gene expression. Science 306:704–708
Oiso N, Fukai K, Kawada A, Suzuki T (2013) Piebaldism. J Dermatol 40:330–335
Pape H (1990) The inheritance of the piebald spotting pattern and its variation in Holstein-Friesian cattle and in Landseer-Newfoundland dogs. Genetica 80:115–128
Quininao C, Prochiantz A, Touboul J (2015) Local homeoprotein diffusion can stabilize boundaries generated by graded positional cues. Development 142:1860–1868
Raspopovic J, Marcon L, Russo L, Sharpe J (2014) Modeling digits. Digit patterning is controlled by a Bmp-Sox9-Wnt Turing network modulated by morphogen gradients. Science 345:566–570
Salsa S, Vegni FMG, Zaretti A, Zunino P (2013) Reaction-diffusion models. In: A primer on PDEs: models, methods, simulations, Springer, Milan, pp 139–188
Sapkota G, Alarcon C, Spagnoli FM, Brivanlou AH, Massague J (2007) Balancing BMP signaling through integrated inputs into the Smad1 linker. Mol Cell 25:441–454
Schneider J (2012) Perfect stripes from a general turing model in different geometries. Boise State University, Boise
Sick S, Reinker S, Timmer J, Schlake T (2006) WNT and DKK determine hair follicle spacing through a reaction–diffusion mechanism. Science 314:1447–1450
Spritz RA (1994) Molecular basis of human piebaldism. J Invest Dermatol 103:137S–140S
Thattai M, van Oudenaarden A (2001) Intrinsic noise in gene regulatory networks. Proc Natl Acad Sci 98:8614–8619
Turing AM (1952) The chemical basis of morphogenesis. Philos Trans R Soc Lond B Biol Sci 237:37–72
Wang Y, Viennet C, Robin S, Berthon JY, He L, Humbert P (2017) Precise role of dermal fibroblasts on melanocyte pigmentation. J Dermatol Sci 88:159–166
Zheng Q, Wang Z, Shen J, Iqbal HMA (2017) Turing bifurcation and pattern formation of stochastic reaction–diffusion system. Adv Math Phys 2017:9
Acknowledgements
The authors wish to thank Yannik Willing, Nina Hedvig Eriksen, and Viktoria Szabolcsi for providing the photographs.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Dougoud, M., Mazza, C., Schwaller, B. et al. Extending the Mathematical Palette for Developmental Pattern Formation: Piebaldism. Bull Math Biol 81, 1461–1478 (2019). https://doi.org/10.1007/s11538-019-00569-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11538-019-00569-1