Abstract
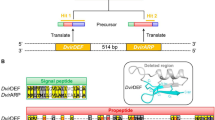
Defensins play a vital role in the early stage of infection before adaptive immune responses are generated. Thus far, only limited detailed genomic data for avian defensin genes have been described. Here, using bacterial artificial chromosome libraries, we found that a 95 kb and 177 kb sequences in the golden pheasant (Chrysolophus pictus, Galliformes) and hwamei (Garrulax canorus, Passeriformes) corresponds to 16 and 30 defensin genes, respectively. Fluorescence in situ hybridization assigned defensin gene clusters in the golden pheasant and hwamei to chromosomes 2q and 3q, respectively. In combination with the previous chicken (Gallus gallus, Galliformes) and zebra finch (Taeniopygia guttata, Passeriformes) defensin clusters, the comparative genomic analysis revealed that lineage-specific duplications and deletions have given rise to clearly different genomic structures. On the basis of genomic characteristics, we further found that transposable elements act as agents of evolution, causing direct and indirect copy number variations in defensin genes via duplications. Tissue expression analysis showed that the Passeriformes-specific duplicated AvBD1 and AvBD3 genes are mainly expressed in the upper and lower respiratory tract, tongue, and spleen. Our analyses indicate that the duplication-and-deletion of defensin genes conformed to the birth-and-death evolutionary process and that transposable elements induced the duplication of defensin genes. Moreover, the respiratory system-specific expression pattern of Passeriformes-specific expanded AvBD1 and AvBD3 genes suggests their important role in maintaining the singing trait of songbirds. The understanding of the genomic structure, expression, and evolution of defensin genes can provide a crucial immunological foundation to study and prevent avian diseases.
摘要
动物先天性免疫系统是病原入侵后遇到的第一道防线,而防御素是先天性免疫系统的重要组成部分。鸟类是传染性病原的重要宿主,其防御素基因的解析有助于阐明鸟类先天性免疫系统的防御机制。然而,有关鸟类防御素基因的研究却鲜见报道。本文解析了红腹锦鸡(鸡形目)和画眉(雀形目)两物种的95和177 kb的防御素基因家族,并结合已有的家鸡(鸡形目)和珍珠鸟(雀形目)防御素基因进行了比较基因组分析,发现鸟类防御素的基因组结构存在世系特异性;鸡形目的红腹锦鸡和家鸡的防御素基因较少,但雀形目的画眉和珍珠鸟的防御素基因却得到了显著膨胀。进一步的组织特异性表达结果表明,雀形目中被特异复制的防御素基因主要表达在上下呼吸道、舌头和脾脏,有助于降低鸣禽呼吸系统的疾病易感性。而且,特异性膨胀的防御素基因特异地表达在鸣禽的呼吸系统,提示防御素基因的膨胀与其鸣唱这一生物特性有关.





Similar content being viewed by others
References
Xiao Y, Hughes AL, Ando J et al (2004) A genome-wide screen identifies a single β-defensin gene cluster in the chicken: implications for the origin and evolution of mammalian defensins. BMC Genom 5:56
van Dijk A, Veldhuizen EJ, Haagsman HP (2008) Avian defensins. Vet Immunol Immunopathol 124:1–18
Yang D, Chertov O, Bykovskaia S et al (1999) β-defensins: linking innate and adaptive immunity through dendritic and t cell ccr6. Science 286:525–528
Hellgren O, Ekblom R (2010) Evolution of a cluster of innate immune genes (β-defensins) along the ancestral lines of chicken and zebra finch. Immunome Res 6:3
Huang Y, Li Y, Burt DW et al (2013) The duck genome and transcriptome provide insight into an avian influenza virus reservoir species. Nat Genet 45:776–783
Morrison GM, Semple CA, Kilanowski FM et al (2003) Signal sequence conservation and mature peptide divergence within subgroups of the murine β-defensin gene family. Mol Biol Evol 20:460–470
Ye Q, He K, Wu S-Y et al (2012) Isolation of a 97-kb minimal essential MHC B locus from a new reverse-4D BAC library of the golden pheasant. PLoS One 7:e32154
Hellgren O, Sheldon B (2011) Locus-specific protocol for nine different innate immune genes (antimicrobial peptides: β-defensins) across passerine bird species reveals within-species coding variation and a case of trans-species polymorphisms. Mol Ecol Res 11:686–692
Wan QH, Zeng CJ, Ni XW et al (2009) Giant panda genomic data provide insight into the birth-and-death process of mammalian major histocompatibility complex class II genes. PLoS One 4:e4147
Liu Y-G, Huang N (1998) Efficient amplification of insert end sequences from bacterial artificial chromosome clones by thermal asymmetric interlaced PCR. Plant Mol Biol Rep 16:175
Wan QH, Zhang P, Ni XW et al (2011) A novel HURRAH protocol reveals high numbers of monomorphic mhc class II loci and two asymmetric multi-locus haplotypes in the père david’s deer. PLoS One 6:e14518
Petersen TN, Brunak S, von Heijne G et al (2011) Signalp 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786
Lynn DJ, Higgs R, Lloyd AT et al (2007) Avian beta-defensin nomenclature: a community proposed update. Immunol Lett 110:86–89
Huang Y, Zhang L (2004) Rapid and sensitive dot-matrix methods for genome analysis. Bioinformatics 20:460–466
Moorhead P, Nowell P, Mellman W et al (1960) Chromosome preparations of leukocytes cultured from human peripheral blood. Exp Cell Res 20:613–616
Wan QH, Pan SK, Hu L et al (2013) Genome analysis and signature discovery for diving and sensory properties of the endangered chinese alligator. Cell Res 23:1091–1105
Tamura K, Dudley J, Nei M et al (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Dereeper A, Guignon V, Blanc G et al (2008) Phylogeny. Fr: robust phylogenetic analysis for the non-specialist. Nucl Acids Res 36:W465–W469
Ronquist F, Huelsenbeck JP (2003) Mrbayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Yang Z (2007) Paml 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24:1586–1591
Weir J, Schluter D (2008) Calibrating the avian molecular clock. Mol Ecol 17:2321–2328
Zhang J (2003) Evolution by gene duplication: an update. Trends Ecol Evol 18:292–298
Marais G, Domazet-Lošo T, Tautz D et al (2004) Correlated evolution of synonymous and nonsynonymous sites in drosophila. J Mol Evol 59:771–779
Cordaux R, Batzer MA (2009) The impact of retrotransposons on human genome evolution. Nat Rev Genet 10:691–703
Quiñones-Mateu ME, Lederman MM, Feng Z et al (2003) Human epithelial beta-defensins 2 and 3 inhibit hiv-1 replication. AIDS 17:F39–F48
van Dijk A, Veldhuizen EJ, Kalkhove SI et al (2007) The β-defensin gallinacin-6 is expressed in the chicken digestive tract and has antimicrobial activity against food-borne pathogens. Antimicrob Agents Chemother 51:912–922
Nei M, Gu X, Sitnikova T (1997) Evolution by the birth-and-death process in multigene families of the vertebrate immune system. Proc Natl Acad Sci USA 94:7799–7806
Schutte BC, Mitros JP, Bartlett JA et al (2002) Discovery of five conserved β-defensin gene clusters using a computational search strategy. Proc Natl Acad Sci USA 99:2129–2133
Swofford DL (2003) PAUP*: phylogenetic analysis using parsimony (* and other methods) (version 4). Sinauer Associates, Sunderland
Felsenstein J (1989) PHYLIP: phylogeny inference package (version 3.2). Cladistics 5:164–166
Fedde M (1998) Relationship of structure and function of the avian respiratory system to disease susceptibility. Poult Sci 77:1130–1138
Davison F, Kaspers B, Schat KA et al (2011) Avian immunology. Academic Press, Massachusetts
Toth TE (2000) Nonspecific cellular defense of the avian respiratory system: a review. Dev Comp Immunol 24:121–139
Hollox EJ, Armour JA, Barber JC (2003) Extensive normal copy number variation of a β-defensin antimicrobial-gene cluster. Am J Hum Genet 73:591–600
Hellgren O, Sheldon B, Buckling A (2010) In vitro tests of natural allelic variation of innate immune genes (avian β-defensins) reveal functional differences in microbial inhibition. J Evol Biol 23:2726–2730
Biémont C, Vieira C (2006) Genetics: junk DNA as an evolutionary force. Nature 443:521–524
Warren WC, Clayton DF, Ellegren H et al (2010) The genome of a songbird. Nature 464:757–762
Hillier LW, Miller W, Birney E et al (2004) Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432:695–716
Kulski JK, Gaudieri S, Bellgard M et al (1997) The evolution of MHC diversity by segmental duplication and transposition of retroelements. J Mol Evol 45:599–609
Völker M, Backström N, Skinner BM et al (2010) Copy number variation, chromosome rearrangement, and their association with recombination during avian evolution. Genome Res 20:503–511
van de Lagemaat LN, Landry J-R, Mager DL et al (2003) Transposable elements in mammals promote regulatory variation and diversification of genes with specialized functions. Trends Genet 19:530–536
Fiston-Lavier A-S, Anxolabehere D, Quesneville H (2007) A model of segmental duplication formation in drosophila melanogaster. Genome Res 17:1458–1470
Acknowledgments
We would like to thank Dr SL Zhang (College of Life Sciences, Zhejiang University) for constructive suggestions for improving the sensitivity of FISH. This work was supported by the National Natural Science Foundation of China (31070334), a special grant from the State Forestry Administration, and the Fundamental Research Funds for the Central Universities of China.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
About this article
Cite this article
Chen, H., Ma, MY., Sun, L. et al. Genomic structure and evolution of beta-defensin genes in the golden pheasant and hwamei. Sci. Bull. 60, 679–690 (2015). https://doi.org/10.1007/s11434-015-0758-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11434-015-0758-3