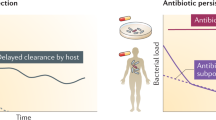
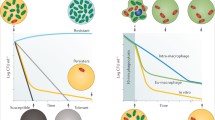
Abstract
Although bacterial persistence was first observed in 1944, its underlying mechanism is just beginning to be understood and many fundamental questions remain. In this review, we summarize studies in order to chart the full map of bacterial persistence. Because persistence significantly contributes to disease recalcitrance, we also elucidate the probable relationships between bacterial persistence and prolonged chronic infections, with some comments on future research directions and therapeutic strategies.
Similar content being viewed by others
References
Lewis K. Persister cells and the riddle of biofilm survival. Biochem J, 2005, 70: 267–274
Bigger J. Treatment of staphylococcal infections with penicillin by intermittent sterilisation. Lancet, 1944, 244: 497–500
Fauvart M, De Groote VN, Michiels J. Role of persister cells in chronic infections: clinical relevance and perspectives on anti-persister therapies. J Med Micobiol, 2011, 60: 699–709
Kint CI, Verstraeten N, Fauvart M, Michiels J. New-found fundamentals of bacterial persistence. Trends Microbiol, 2012, 20: 577–585
Jayaraman R. Bacterial persistence: some new insights into an old phenomenon. J Biosci, 2008, 33: 795–805
Cohen NR, Lobritz MA, Collins JJ. Microbial persistence and the road to drug resistance. Cell Host Microbe, 2013, 13: 632–642
Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial persistence as a phenotypic switch. Science, 2004, 305: 1622–1625
Fung DK, Chan EW, Chin ML, Chan RC. Delineation of a bacterial starvation stress response network which can mediate antibiotic tolerance development. Agent Chemother, 2010, 54: 1082–1093
Maisonneuve E, Shakespeare LJ, Jørgensen MG, Gerdes K. Bacterial persistence by RNA endonucleases. Pro Nat Acad Sci USA, 2011, 108: 13206–13211
Maisonneuve E, Castro-Camargo M, Gerdes K. (p)ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell, 2013, 154: 1140–1150
Lee HH, Molla MN, Cantor CR, Collins JJ. Bacterial charity work leads to population-wide resistance. Nature, 2010, 467: 82–85
Wakamoto Y, Dhar N, Chait R, Schneider K, Signorino-Gelo F, Leibler S, McKinney JD. Dynamic persistence of antibiotic-stressed mycobacteria. Science, 2013, 339: 91–95
Delvigne F, Goffin P. Microbial heterogeneity affects bioprocess robustness: dynamic single-cell analysis contributes to understanding of microbial populations. Biotechnol J, 2014, 9: 61–72
Theodore A, Lewis K, Vulić M. Tolerance of Escherichia coli to fluoroquinolone antibiotics depends on specific components of the SOS response pathway. Genetics, 2013, 195: 1265–1276
Kwan BW, Valenta JA, Benedik MJ, Wood TK. Arrested protein synthesis increases persister-like cell formation. Agent Chemother, 2013, 57: 1468–1473
Amato SM, Orman MA, Brynildsen MP. Metabolic control of persister formation in Escherichia coli. Mol Cell, 2013, 50: 475–487
Vega NM, Allison KR, Khalil AS, Collins JJ. Signaling-mediated bacterial persister formation. Nat Chem Biol, 2012, 8: 431–433
Hayes F. Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science, 2003, 301: 1496–1499
Lewis K. Persister cells, dormancy and infectious disease. Nat Rev Microb, 2007, 5: 48–56
Kostakioti M, Hadjifrangiskou M, Hultgren SJ. Bacterial biofilms: development, dispersal, and therapeutic strategies in the dawn of the postantibiotic era. Cold Spring Harb Perspect Med, 2013, 3: a010306
Engelberg-Kulka H, Amitai S, Kolodkin-Gal I, Hazan R. Bacterial programmed cell death and multicellular behavior in bacteria. PLoS Genet, 2006, 2: e135
Tripathi A, Dewan PC, Siddique SA, Varadarajan R. MazF induced growth inhibition and persister generation in Escherichia coli. J Bio Chem, 2014, 289: 4191–4205
Hong SH, Wang X, O’Connor HF, Benedik MJ, Wood TK. Bacterial persistence increases as environmental fitness decreases. Microb Biotechnol, 2012, 5: 509–522
Wayne LG, Hayes LG. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect Immun, 1996, 64: 2062–2069
Wu Y, Vulić M, Keren I, Lewis K. Role of oxidative stress in persister tolerance. Agent Chemother, 2012, 56: 4922–4926
Foti JJ, Devadoss B, Winkler JA, Collins JJ, Walker GC. Oxidation of the guanine nucleotide pool underlies cell death by bactericidal antibiotics. Science, 2012, 336: 315–319
Gusarov I, Shatalin K, Starodubtseva M, Nudler E. Endogenous nitric oxide protects bacteria against a wide spectrum of antibiotics. Science, 2009, 325: 1380–1384
Shatalin K, Shatalina E, Mironov A, Nudler E. H2S: a universal defense against antibiotics in bacteria. Science, 2011, 334: 986–990
Anderson KL, Whitlock JE, Harwood VJ. Persistence and differential survival of fecal indicator bacteria in subtropical waters and sediments. Appl Environ Microbiol, 2005, 71: 3041–3048
Leung V, Lévesque CM. A stress-inducible quorum-sensing peptide mediates the formation of persister cells with noninherited multidrug tolerance. J Bacteriol, 2012, 194: 2265–2274
Park SJ, Son WS, Lee BJ. Structural overview of toxin-antitoxin systems in infectious bacteria: a target for developing antimicrobial agents. BBA-Proteins Proteom, 2013, 1834: 1155–1167
Hayes F, Kędzierska B. Regulating toxin-antitoxin expression: controlled detonation of intracellular molecular timebombs. Toxins, 2014, 6: 337–358
Lusetti SL, Cox MM. The bacterial RecA protein and the recombinational DNA repair of stalled replication forks. Annu Rev Biochem, 2002, 71: 71–100
Radman M. SOS repair hypothesis: phenomenology of an inducible DNA repair which is accompanied by mutagenesis. In: Hanawalt P, Eds. Basic Life Sciences. Vol. 5: Molecular Mechanisms for Repair of DNA. New York: Springer US, 1975. 355–367
Sassanfar M, Roberts JW. Nature of the sos-inducing signal in Escherichia coli: the involvement of DNA replication. J Mol Biol, 1990, 212: 79–96
Kimmitt PT, Harwood CR, Barer MR. Toxin gene expression by shiga toxin-producing Escherichia coli: the role of antibiotics and the bacterial SOS response. Emerg Infect Dis, 2000, 6: 458–465
Guerin É, Cambray G, Sanchez-Alberola N, Campoy S, Erill I, Da ReS, Gonzalez-Zorn B, Barbé J, Ploy MC, Mazel D. The SOS response controls integron recombination. Science, 2009, 324: 1034–1034
Kolter R, Greenberg EP. Microbial sciences: the superficial life of microbes. Nature, 2006, 441: 300–302
Möker N, Dean CR, Tao J. Pseudomonas aeruginosa increases formation of multidrug-tolerant persister cells in response to quorum-sensing signaling molecules. J Bacteriol, 2010, 192: 1946–1955
Rutherford ST, Bassler BL. Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb Perspect Med, 2012, 2: a012427
Jain V, Kumar M, Chatterji D. ppGpp: stringent response and survival. J Microbiol, 2006, 44: 1–10
Engelberg-Kulka H, Glaser G. Addiction modules and programmed cell death and antideath in bacterial cultures. Annu Rev Microbiol, 1999, 53: 43–70
Drlica K. Control of bacterial DNA supercoiling. Mol Microbiol, 1992, 6: 425–433
Pruss GJ, Manes SH, Drlica K. Escherichia coli DNA topoisomerase I mutants: increased supercoiling is corrected by mutations near gyrase genes. Cell, 1982, 31: 35–42
Ryan RP. Cyclic di-GMP signalling and the regulation of bacterial virulence. Microbiol, 2013, 159: 1286–1297
Krasteva PV, Giglio KM, Sondermann H. Sensing the messenger: the diverse ways that bacteria signal through c-di-GMP. Protein Sci, 2012, 21: 929–948
Kalia D, Merey G, Nakayama S, Zheng Y, Zhou J, Luo Y, Guo M, Roembke BT, Sintim HO. Nucleotide, c-di-GMP, c-di-AMP, cGMP, cAMP, (p)ppGpp signaling in bacteria and implications in pathogenesis. Chem Soc Rev, 2013, 42: 305–341
Chimerel C, Field CM, Piñero-Fernandez S, Keyser UF, Summers DK. Indole prevents Escherichia coli cell division by modulating membrane potential. BBA-Biomembranes, 2012, 1818: 1590–1594
Li XZ, Ma D, Livermore DM, Nikaido H. Role of efflux pump(s) in intrinsic resistance of Pseudomonas aeruginosa: active efflux as a contributing factor to beta-lactam resistance. Agent Chemother, 1994, 38: 1742–1752
Jones SE, Lennon JT. Dormancy contributes to the maintenance of microbial diversity. Proc Nat Acad Sci USA, 2010, 107: 5881–5886
Ferullo DJ, Lovett ST. The stringent response and cell cycle arrest in Escherichia coli. PLoS Genet, 2008, 4: e1000300
Garnett JA, Matthews S. Interactions in bacterial biofilm development: a structural perspective. Curr Protein Pept Sci, 2012, 13: 739–755
Lewis K. Riddle of biofilm resistance. Agent Chemother, 2001, 45: 999–1007
Adams KN, Takaki K, Connolly LE, Wiedenhoft H, Winglee K, Humbert O, Edelstein PH, Cosma CL, Ramakrishnan L. Drug tolerance in replicating mycobacteria mediated by a macrophage-induced efflux mechanism. Cell, 2011, 145: 39–53
Schaeffer P, Millet J, Aubert JP. Catabolic repression of bacterial sporulation. Proc Nat Acad Sci USA, 1965, 54: 704–711
Grant SS, Kaufmann BB, Chand NS, Haseley N, Hung DT. Eradication of bacterial persisters with antibiotic-generated hydroxyl radicals. Proc Nat Acad Sci USA, 2012, 109: 12147–12152
Conlon B, Nakayasu E, Fleck LE, LaFleur MD, Isabella VM, Coleman K, Leonard SN, Smith RD, Adkins JN, Lewis K. Activated ClpP kills persisters and eradicates a chronic biofilm infection. Nature, 2013, 503: 365–370
Allison KR, Brynildsen MP, Collins JJ. Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature, 2011, 473: 216–220
Fu Y, Zhu M, Xing J. Resonant activation: a strategy against bacterial persistence. Phys Biol, 2010, 7: 016013
Veening JW, Smits WK, Kuipers OP. Bistability, epigenetics, and bet-hedging in bacteria. Annu Rev Microbiol, 2008, 62: 193–210
Spudich JL, Koshland Jr D. Non-genetic individuality: chance in the single cell. Nature, 1976, 262: 467–471
Mora T, Bai F, Che YS, Minamino T, Namba K, Wingreen NS. Non-genetic individuality in Escherichia coli motor switching. Phys Biol, 2011, 8: 024001
Bai F, Che YS, Kami-ike N, Ma Q, Minamino T, Sowa Y, Namba K. Populational heterogeneity vs. temporal fluctuation in Escherichia coli flagellar motor switching. Biophys J, 2013, 105: 2123–2129
Iino R, Matsumoto Y, Nishino K, Yamaguchi A, Noji H. Design of a large-scale femtoliter droplet array for single-cell analysis of drug-tolerant and drug-resistant bacteria. Front Microbiol, 2013, 4: 300
Taniguchi Y, Choi PJ, Li G-W, Chen H, Babu M, Hearn J, Emili A, Xie XS. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science, 2010, 329: 533–538
Author information
Authors and Affiliations
Corresponding authors
Additional information
These authors contributed equally to this work
Rights and permissions
About this article
Cite this article
Liu, Y., Yang, J., Zhao, Z. et al. Bacterial persistence. Sci. China Chem. 57, 1625–1633 (2014). https://doi.org/10.1007/s11426-014-5245-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11426-014-5245-1