Abstract
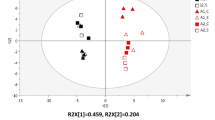
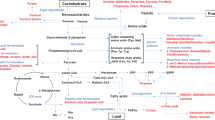
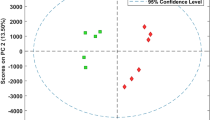
New approaches, mainly based on mass spectrometry techniques, are being developed and appear as a must in the modern food science and microbiology research to investigate food quality and safety. To date, the investigation of cheese ripening mechanisms has mostly used targeted approaches. The aims of the present project were to assess the use of untargeted metabolomics as an approach to investigate the influence of altering one ripening parameter to generate fine differences in the microbial metabolism within cheese. Two cheeses were made which varied with respect to the spatial distribution of bacterial colonies, leading to cheeses with only big or only small colonies. Liquid chromatography high resolution mass spectrometry metabolic fingerprints were acquired on cheese extracts collected after 2, 13 and 27 days of ripening using two different extraction methods (water or acetonitrile) and analyzed using two different simultaneous ionization modes (positive and negative electrospray). Data processing involving XCMS and multivariate statistical analysis highlighted significant discriminant profiles of the cheese metabolomes according to the two different spatial distributions compared. The different fractions investigated (water and acetonitrile extractions in two ionization modes) were complementary and resulted in a view as global as possible of the cheese metabolome which had been modulated by the spatial distribution of bacterial colonies. Some of the metabolites were then identified using an in-house database. These results show the relevance of cheese LC–HRMS fingerprinting to understand the influence of a ripening parameter generating fine differences on microbial metabolism within cheese.




Similar content being viewed by others
References
Aly, S., Floury, J., Famelart, M. H., Madec, M. N., Dupont, D., Le Gouar, Y., et al. (2011). Nisin quantification by ELISA qllows the modeling of its apparent diffusion coefficient in model cheeses. Journal of Agricultural and Food Chemistry, 59(17), 9484–9490.
Antignac, J., Courant, F., Pinel, G., Bichon, E., Monteau, F., Elliott, C., et al. (2011). Mass spectrometry-based metabolomics applied to the chemical safety of food. Trends in Analytical Chemistry, 30(2), 292–301.
Buscher, J. M., Czernik, D., Ewald, J. C., Sauer, U., & Zamboni, N. (2009). Cross-platform comparison of methods for quantitative metabolomics of primary metabolism. Analytical Chemistry, 81(6), 2135–2143.
Cevallos-Cevallos, J. M., Reyes-De-Corcuera, J. I., Etxeberria, E., Danyluk, M. D., & Rodrick, G. E. (2009). Metabolomic analysis in food science: A review. Trends in Food Science & Technology, 20(11-12), 557–566.
Cifuentes, A. (2009). Food analysis and foodomics foreword. Journal of Chromatography A, 1216(43), 7109.
Consonni, R., & Cagliani, L. (2008). Ripening and geographical characterization of Parmigiano Reggiano cheese by (1)H NMR spectroscopy. Talanta, 76(1), 200–205.
Courant, F., Royer, A. L., Chéreau, S., Morvan, M., Monteau, F., Antignac, J. P., et al. (2012). Implementation of a semi-automated strategy for the annotation of metabolomic fingerprints generated by liquid chromatography-high resolution mass spectrometry from biological samples. Analyst, 137(21), 4958–4967.
Cretenet, M., Laroute, V., Ulve, V., Jeanson, S., Nouaille, S., Even, S., et al. (2011). Dynamic analysis of the Lactococcus lactis transcriptome in cheeses made from milk concentrated by ultrafiltration reveals multiple strategies of adaptation to stresses. Applied and Environmental Microbiology, 77(1), 247–257.
Dunn, W. B., & Ellis, D. I. (2005). Metabolomics: Current analytical platforms and methodologies. Trac-Trends in Analytical Chemistry, 24(4), 285–294.
Duportet, X., Aggio, R. B. M., Carneiro, S., & Villas-Boas, S. G. (2012). The biological interpretation of metabolomic data can be misled by the extraction method used. Metabolomics, 8(3), 410–421.
Garcia-Canas, V., Simo, C., Herrero, M., Ibanez, E., & Cifuentes, A. (2012). Present and future challenges in food analysis: Foodomics. Analytical Chemistry, 84(23), 10150–10159.
Gianferri, R., Maioli, M., Delfini, M., & Brosio, E. (2007). A low-resolution and high-resolution nuclear magnetic resonance integrated approach to investigate the physical structure and metabolic profile of Mozzarella di Bufala Campana cheese. International Dairy Journal, 17(2), 167–176.
Goodacre, R., Vaidyanathan, S., Dunn, W. B., Harrigan, G. G., & Kell, D. B. (2004). Metabolomics by numbers: Acquiring and understanding global metabolite data. Trends in Biotechnology, 22(5), 245–252.
Hannon, J. A., Lopez, C., Madec, M. N., & Lortal, S. (2006). Altering renneting pH changes microstructure, cell distribution, and lysis of Lactococcus lactis AM2 in cheese made from ultrafiltered milk. Journal of Dairy Science, 89(3), 812–823.
Hendriks, M. M. W. B., van Eeuwijk, F. A., Jellema, R. H., Westerhuis, J. A., Reijmers, T. H., Hoefsloot, H. C. J., et al. (2011). Data-processing strategies for metabolomics studies. Trac-Trends in Analytical Chemistry, 30(10), 1685–1698.
Herrero, M., Simo, C., Garcia-Canas, V., Ibanez, E., & Cifuentes, A. (2012). Foodomics: MS-based strategies in modern food science and nutrition. Mass Spectrometry Reviews, 31(1), 49–69.
Hession, A. O., Esrey, E. G., Croes, R. A., & Maxwell, C. A. (2008). N-acetylglutamate and N-acetylaspartate in soybeans (Glycine max L.), maize (Zea maize L.), and other foodstuffs. Journal of Agricultural and Food Chemistry, 56(19), 9121–9126.
Isolini, D., Grand, M., & Glattli, H. (1990). Selective media for the detection of obligate and facultative heterofermentative lactobacilli. Schweizerische Milchwirtschaftliche Forschung, 19(3), 57–59.
Jeanson, S., Chadoeuf, J., Madec, M., Aly, S., Floury, J., Brocklehurst, T., et al. (2011). Spatial distribution of bacterial colonies in a model cheese. Applied and Environmental Microbiology, 77(4), 1493–1500.
Juillard, V., Lebars, D., Kunji, E. R. S., Konings, W. N., Gripon, J. C., & Richard, J. (1995). Oligopeptides are the main source of nitrogen for Lactococcus lactis during growth in milk. Applied and Environmental Microbiology, 61(8), 3024–3030.
Kell, D. B., & Oliver, S. G. (2004). Here is the evidence, now what is the hypothesis? The complementary roles of inductive and hypothesis-driven science in the post-genomic era. BioEssays, 26(1), 99–105.
Kessner, D., Chambers, M., Burke, R., Agusand, D., & Mallick, P. (2008). ProteoWizard: Open source software for rapid proteomics tools development. Bioinformatics, 24(21), 2534–2536.
Kilstrup, M., Hammer, K., Jensen, P. R., & Martinussen, J. (2005). Nucleotide metabolism and its control in lactic acid bacteria. FEMS Microbiology Reviews, 29(3), 555–590.
Le Boucher, C., Courant, F., Jeanson, S., Chereau, S., Maillard, M. B., Royer, A. L., et al. (2013). First mass spectrometry metabolic fingerprinting of bacterial metabolism in a model cheese. Food Chemistry, 141(2), 1032–1040.
McSweeney, P. L. H. (2004). Biochemistry of cheese ripening. International Journal of Dairy Technology, 57(2-3), 127–144.
Meldrum, R. J., Brocklehurst, T. F., Wilson, D. R., & Wilson, P. D. G. (2003). The effects of cell immobilization, pH and sucrose on the growth of Listeria monocytogenes Scott A at 10 degrees C. Food Microbiology, 20(1), 97–103.
Mucchetti, G., Locci, F., Gatti, M., Neviani, E., Addeo, F., Dossena, A., et al. (2000). Pyroglutamic acid in cheese: Presence, origin, and correlation with ripening time of Grana Padano cheese. Journal of Dairy Science, 83(4), 659–665.
Neumann, S., & Bocker, S. (2010). Computational mass spectrometry for metabolomics: Identification of metabolites and small molecules. Analytical and Bioanalytical Chemistry, 398(7-8), 2779–2788.
Niven, G. W., Knight, D. J., & Mulholland, F. (1998). Changes in the concentrations of free amino acids in milk during growth of Lactococcus lactis indicate biphasic nitrogen metabolism. Journal of Dairy Research, 65(1), 101–107.
Patti, G. J., Yanes, O., & Siuzdak, G. (2012). Metabolomics: the apogee of the omics trilogy. Nature Reviews Molecular Cell Biology, 13(4), 263–269.
Preininger, M., Warmke, R., & Grosch, W. (1996). Identification of the character impact flavour compounds of Swiss cheese by sensory studies of models. Zeitschrift fur Lebensmittel-Untersuchung Und-Forschung, 202(1), 30–34.
Putri, S. P., Nakayama, Y., Matsuda, F., Uchikata, T., Kobayashi, S., Matsubara, A., et al. (2013). Current metabolomics: Practical applications. Journal of Bioscience and Bioengineering, 115(6), 579–589.
Ryan, D., & Robards, K. (2006). Metabolomics: The greatest omics of them all? Analytical Chemistry, 78(23), 7954–7958.
Shintu, L., & Caldarelli, S. (2006). Toward the determination of the geographical origin of emmental(er) cheese via high resolution MAS NMR: A preliminary investigation. Journal of Agricultural and Food Chemistry, 54(12), 4148–4154.
Skandamis, P., Tsigarida, E., & Nychas, G. J. E. (2000). Ecophysiological attributes of Salmonella typhimurium in liquid culture and within a gelatin gel with or without the addition of oregano essential oil. World Journal of Microbiology & Biotechnology, 16(1), 31–35.
Smith, C. A., Want, E. J., O’Maille, G., Abagyan, R., & Siuzdak, G. (2006). XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Analytical Chemistry, 78(3), 779–787.
Song, A. A. L., Abdullah, J. O., Abdullah, M. P., Shafee, N., Othman, R., Tan, E. F., et al. (2012). Overexpressing 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMGR) in the lactococcal mevalonate pathway for heterologous plant sesquiterpene production. Plos One, 7(12), e52444.
Sumner, L. W., Amberg, A., Barrett, D., Beale, M. H., Beger, R., Daykin, C. A., et al. (2007). Proposed minimum reporting standards for chemical analysis. Metabolomics, 3(3), 211–221.
Tautenhahn, R., Patti, G. J., Kalisiak, E., Miyamoto, T., Schmidt, M., Lo, F. Y., et al. (2011). metaXCMS: Second-order analysis of untargeted metabolomics data. Analytical Chemistry, 83(3), 696–700.
Theodoridis, G., Gika, H. G., & Wilson, I. D. (2011). Mass spectrometry-based holistic analytical approaches for metabolite profiling in systems biology studies. Mass Spectrometry Reviews, 30(5), 884–906.
Ulve, V., Monnet, C., Valence, F., Fauquant, J., Falentin, H., & Lortal, S. (2008). RNA extraction from cheese for analysis of in situ gene expression of Lactococcus lactis. Journal of Applied Microbiology, 105(5), 1327–1333.
van der Kloet, F. M., Bobeldijk, I., Verheij, E. R., & Jellema, R. H. (2009). Analytical error reduction using single point calibration for accurate and precise metabolomic phenotyping. Journal of Proteome Research, 8(11), 5132–5141.
Villas-Boas, S. G., Hojer-Pedersen, J., Akesson, M., Smedsgaard, J., & Nielsen, J. (2005). Global metabolite analysis of yeast: Evaluation of sample preparation methods. Yeast, 22(14), 1155–1169.
Walker, S. L., Brocklehurst, T. F., & Wimpenny, J. W. T. (1997). The effects of growth dynamics upon pH gradient formation within and around subsurface colonies of Salmonella typhimurium. Journal of Applied Microbiology, 82(5), 610–614.
Wilson, P. D. G., Brocklehurst, T. F., Arino, S., Thuault, D., Jakobsen, M., Lange, M., et al. (2002). Modelling microbial growth in structured foods: Towards a unified approach. International Journal of Food Microbiology, 73(2-3), 275–289.
Winder, C. L., Dunn, W. B., Schuler, S., Broadhurst, D., Jarvis, R., Stephens, G. M., et al. (2008). Global metabolic profiling of Escherichia coli cultures: An evaluation of methods for quenching and extraction of intracellular metabolites. Analytical Chemistry, 80(8), 2939–2948.
Wishart, D. S. (2008). Metabolomics: Applications to food science and nutrition research. Trends in Food Science & Technology, 19(9), 482–493.
Yanes, O., Tautenhahn, R., Patti, G. J., & Siuzdak, G. (2011). Expanding coverage of the metabolome for global metabolite profiling. Analytical Chemistry, 83(6), 2152–2161.
Yvon, M., & Rijnen, L. (2001). Cheese flavour formation by amino acid catabolism. International Dairy Journal, 11(4-7), 185–201.
Acknowledgments
This work was performed in the framework of the CheeseOmic Project co-funded by the Brittany and Pays-de-la-Loire Regional Councils (France) and supported by Bretagne Biotechnologie Alimentaire (Bba) association. Clémentine Le Boucher is recipient of a PhD Grant from the French Ministry of Research. Authors are grateful to Marie-Bernadette Maillard for her support concerning microbial analysis and to Hector Gallart-Ayala for his support concerning the data analysis.
Conflict of interest
The authors declare no conflict of interests.
Compliance with ethical requirements
This article does not contain any studies with human or animal subjects.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Le Boucher, C., Courant, F., Royer, AL. et al. LC–HRMS fingerprinting as an efficient approach to highlight fine differences in cheese metabolome during ripening. Metabolomics 11, 1117–1130 (2015). https://doi.org/10.1007/s11306-014-0769-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11306-014-0769-0