Abstract
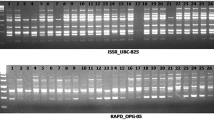
Genetic diversity of Pongamia pinnata, a plant species emerging as biodiesel source, was assessed using three endonuclease (TE)-amplified fragment length polymorphism (AFLP) markers. A total of 275 accessions from different Indian geographical regions were collected. High level of polymorphism (62.6%) was observed in 334 loci amplified using six primer combinations. The Jaccard’s dissimilarity coefficient ranged from 0.17 to 0.80. Based on the neighbour joining method of clustering, accessions were grouped into six major clusters congruent with their respective geographical locations. The mean values of effective multiplex ratio, polymorphic information content, marker index and resolving power were 34.83, 0.286, 9.700 and 14.29 respectively. In addition, a core collection consisting of 20 accessions was identified. Population STRUCTURE analysis produced the highest log likelihood score when the number of populations (K) was set at 2, which was not consistent with clustering analysis based on genetic diversity. Two TE-AFLP markers, E_ac x P_a04 and E_ac x P_a39, were significantly associated with oil content (at P = 0.005). The extent of genetic diversity detected and core germplasm of CPT identified in this study could be selected and used for tree breeding, germplasm conservation and suitable feed stock for biofuel.




Similar content being viewed by others
References
Azam MM, Waris A, Nahar NM (2005) Prospects and potential of fatty acid methyl esters of some non-traditional seed oils for use as biodiesel in India. Biomass Bioenerg 29:293–302
Dent A,E, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Evanno G, Regnout S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Falush D, Stephens M, Pritchard J (2003) Inference of Population Structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Falush D, Stephens M, Pritchard JK (2007) Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes 7:574–578
Jaccard P (1908) Nouvelles rescherches sur la distribution florale. Bulletin de la Société vaudoise des sciences naturelles 44:223–270
Karmee SK, Chadha A (2005) Preparation of biodiesel from crude oil of Pongamia pinnata. Bioresour Technol 96:1425–1429
Kaushik N, Kumar S, Kumar K, Beniwal R, Kaushik N, Roy S (2007) Genetic variability and association studies in pod and seed traits of Pongamia pinnata (L.) Pierre in Haryana, India. Genet Res Crop Evol 54:1827–1832
Kesari V, Krishnamachari A, Rangan L (2008) Systematic characterisation and seed oil analysis in candidate plus trees of biodiesel plant, Pongamia pinnata. Ann Appl Biol 152:397–404
Kesari V, Madurai V, Sathyanarayana, Parida A, Rangan L (2010) Molecular marker-based characterization in candidate plus trees of Pongamia pinnata, a potential biodiesel legume. AoB Plants. doi:10.1093/aobpla/plq017
Kim K-W, Chung H-K, Cho G-T, Ma K-H, Chandrabalan D, Gwag J-G, Kim T-S, Cho E-G, Park Y-J (2007) PowerCore: a program applying the advanced M strategy with a heuristic search for establishing core sets. Bioinformatics 23:2155–2162
Krutovsky KV, Clair JBS, Saich R, Hipkins VD, Neale DB (2009) Estimation of population structure in coastal Douglas-fir Pseudotsuga menziesii (Mirb.) Franco var. menziesii using allozyme and microsatellite markers. Tree Genet Genomes 5:641–658
Lakshmikanthan V (1978) Tree borne oil seeds. In: Directorate of non edible oils and soap industries. Khadi and Village Industries Commission, Mumbai, India, p. 10
Mukta N, Murthy IYLN, Sripal P (2009) Variability assessment in Pongamia pinnata (L.) Pierre germplasm for biodiesel traits. Ind Crop Prod 29:536–540
Pavithra HR, Shivanna MB, Chandrika K, Prasanna KT, Gowda B (2014) Genetic analysis of Pongamia pinnata (L.) Pierre populations using AFLP markers. Tree Genet Genomes 10:173–188
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Perrier X, Jacquemoud-Collet JP (2006) DARwin software, Version 5.0.158. http://darwin.cirad.fr/darwin
Powell W, Morgante M, Andre C, Hanafey M, Vogel J, Tingey S, Rafalski A (1996) The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol Breed 2:225–238
Prevost A, Wilkinson MJ (1999) A new system of comparing PCR primers applied to ISSR fingerprinting of potato cultivars. Theor Appl Genet 98:107–112
Pritchard J, Stephens M, Rosenberg N, Donnelly P (2000) Association mapping in structured populations. Am J Hum Genet 67(170):181
Roldan-Ruiz I, Dendauw J, VanBockstaele E, Depicker A, Loose MD (2000) AFLP markers reveal high polymorphic rates in ryegrasses (Lolium spp.). Mol Breed 6:125–134
Sahoo D, Aparajita S, Rout G (2010) Inter and intra-population variability of Pongamia pinnata: a bioenergy legume tree. Plant Sys Evol 285:121–125
Scott PT, Pregelj L, Chen N, Hadler JS, Djordjevic MA, Gresshoff PM (2008) Pongamia pinnata: an untapped resource for the biofuels industry of the future. Bioenerg Res 1:2–11
Sharma RK, Negi MS, Sharma S, Bhardwaj P, Kumar R, Bhattacharya E, Tripathi SB, Vijayan D, Baruah AR, Das SC, Bera B, Rajkumar R, Thomas J, Sud RK, Muraleedharan N, Hazarika M, Lakshmikumaran M, Raina SN, Ahuja PS (2010) AFLP-based genetic diversity assessment of commercially important tea germplasm in India. Biochem Genet 48:549–564
Sharma SS, Aadil K, Negi MS, TripathI SB (2014) Efficacy of two dominant marker systems, ISSR and TE-AFLP for assessment of genetic diversity in biodiesel species Pongamia pinnata. Curr Sci 106:1576–1580
Sharma SS, Islam MA, Malik AA, Kumar K, Negi MS, Tripathi SB (2016) Seed traits, fatty acid profile and genetic diversity assessment in Pongmia pinnata (L.) Pierre germplasm. Physiol Mol Biol Plants. doi:10.1007/s12298-016-0356-0
Sharma SS, Negi MS, Sinha P, Kumar K, Tripathi SB (2011) Assessment of genetic diversity of biodiesel species Pongamia pinnata accessions using AFLP and three endonuclease–AFLP. Plant Mol Biol Rep 29:12–18
Singh A, Negi MS, Rajagopal J, Bhatia S, Tomar UK, Srivastava PS, Lakshmikumaran M (1999) Assessment of genetic diversity in Azadirachta indica using AFLP markers. Theor Appl Genet 99:272–279
Sinha P, Negi MS, Sharma SS, Md Islam A, Tripathi SB (2013) Analysis of genome-wide homozygosity in Jatropha curcas accessions using AFLP markers. Int J Res Pharm Sci 3:191–201
Tewari DN (2003) Report of the committee on development of bio-fuel. Planning Commission, Government of India
Thudi M, Revathi M, Wani SP, Tatikonda L, Hoisington DA, Varshney RK (2010) Analysis of genetic diversity in Pongamia [Pongamia pinnata (L) Pierre] using AFLP markers. J Plant Biochem Biotech 19:209–216
van der Wurff AWG, Chan YL, van Straalen NM, Schouten J (2000) TE-AFLP: combining rapidity and robustness in DNA fingerprinting. Nucl Acids Res 28:e105
Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Friters A, Pot J, Paleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucl Acids Res 23:4407–4414
Acknowledgements
The authors sincerely acknowledge the funding received from the Department of Science and Technology, Government of India, for the project (grant number SR/SO/PS-08/2006). Mr. Shyam Sundar Sharma thanks the Council of Scientific and Industrial Research, Government of India, for providing fellowship for this work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Data archiving statement
The data consisting of raw data and primary information of 275 individuals will be archived in Dryad (http://datadryad.org/).
Additional information
Responsible editor: J. L. Wegrzyn
Electronic supplementary material
ESM 1
List of Pongamia accessions used in the study (DOC 289 kb)
ESM 2
(A) Map showing the geographical locations of collection sites of Pongamia accessions used in this study. The four red dots represent accessions from different regions of Delhi, two orange dots represent East and South NCR. (B) The yellow dots showing the collection sites in other states of India (DOC 113 kb)
ESM 3
Representative autoradiogram of TE-AFLP fingerprints of Pongamia pinnata accessions using E-AG x P-C primer combination (DOC 976 kb)
Rights and permissions
About this article
Cite this article
Sharma, S.S., Islam, M.A., Singh, V.K. et al. Genetic diversity, population structure and association study using TE-AFLP markers in Pongamia pinnata (L.) Pierre germplasm. Tree Genetics & Genomes 13, 6 (2017). https://doi.org/10.1007/s11295-016-1088-6
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-016-1088-6