Abstract
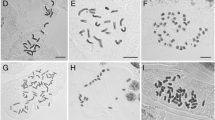
Polyploidization is a major source of diversification among plants, particularly during cladogenesis, but most evidence involves herbaceous temperate species. The prevalence of polyploidy among woody taxa is largely unknown, especially among tropical groups. In this study, we examined genome size variation globally and at several taxonomic levels within the Fagaceae. This family has diversified in the northern temperate zone (Quercus) and at least twice in the Asian tropics (Lithocarpus and Castanopsis), allowing us to examine genomic size evolution across a broad latitudinal range. We compared nuclear DNA contents from 78 species in six genera, including new measurements for 171 individuals from 47 Chinese species using standard flow cytometry methods. No evidence suggests that polyploidization or whole genome duplication has occurred in the family. Genome size varied among genera, but limited variation was present in each genus and species. In general, tropical species had larger genomes than temperate species, but the ancestral state cannot be determined given current evidence. Partial duplication does seem to occur among species as within genus variation was larger than within species variation. A review of the literature suggests that genome size and even chromosome structure is highly conserved among woody plants and trees. We propose that ploidy level and genome size are conserved among trees because they participate in diverse syngameons. This behavior would provide similar benefits to polyploidization but avoid exclusion from the syngameon. This conservatism in genome size and structure should enhance ongoing whole genome studies.



Similar content being viewed by others
References
Adams KL, Wendel JF (2005) Polyploidy and genome evolution in plants. Curr Opin Plant Biol 8:135–141
Armstron JM, Wylie AP (1965) A new basic chromosome number in the family Fagaceae. Nature 205:1340–1341
Arnold ML, Bouck AC, Scott Cornman R (2003) Verne Grant and Louisiana Irises: Is there anything new under the sun? Research review. New Phytol 161:143–149. doi:10.1046/j.1469-8137.2003.00856.x
Beaulieu JM, Smith SA, Leitch IJ (2010) On the tempo of genome size evolution in angiosperms. J Bot. doi:10.1155/2010/989152
Bennett MD (1972) Nuclear DNA content and minimum generation time in herbaceous plants. Proc R Soc Lond Ser B Biol Sci 181:109–135
Bennett MD (1987) Variation in genomic form in plants and its ecological implications. New Phytol 106:177–200
Bennett MD (1998) Plant genome values: How much do we know? Proc Natl Acad Sci U S A 95:2011–2016
Bennett MD, Leitch IJ (2010) Angiosperm DNA C-values Database (release 7.0, Dec. 2010). http://www.kew.org/cvalues/. Accessed 19 Jul. 2011
Bennett MD, Leitch IJ (2011) Nuclear DNA amounts in angiosperms: targets, trends and tomorrow. Ann Bot 107:467–590
Bennetzen JL, Kellogg EA (1997) Do plants have a one-way ticket to genomic obesity? Plant Cell 9:1509–1514
Bowers JE, Chapman BA, Rong J, Paterson AH (2003) Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422:433–438. doi:10.1038/nature01521
Camus A (1934–1954) Les chenes monographie du genre Quercus (et Lithocarpus). Encyclopedie economique de sylviculture, vol 6-8. Academie des Sciences, Paris
Chen G, Sun WB (2010) Ploidy variation in Trigonobalanus verticillata (Fagaceae). Plant Syst Evol 284:123–127
Chen G, Sun WB, Han CY, Coombes A (2007) Karyomorphology of the endangered Trigonobalanus doichangensis (A. Camus) Forman (Fagaceae) and its taxonomic and biogeographical implications. Bot J Linn Soc 154:321–330
Chokchaichamnankit P, Chulalaksananukul W, Phengklai C, Anamthawat-Jonsson K (2007) Karyotypes of some species of Castanopsis, Lithocarpus and Quercus (Fagaceae) from Khun Mae Kuang Forest in Chiang Mai province, northern Thailand. Thai For Bull (Botany) 35:38–44
Crepet WL, Nixon KC (1989) Earliest megafossil evidence of Fagaceae: phylogenetic and biogeographic implications. Am J Bot 76:842–855
Development Core Team R (2010) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Dickson EE, Arumuganathan K, Kresovich S, Doyle JJ (1992) Nuclear DNA Content Variation within the Rosaceae. Am J Bot 79:1081–1086
Doležel J, Greilhuber J, Lucretti S, Meister A, Lysak MA, Nardi L, Obermayer R (1998) Plant genome size estimation by flow cytometry: Inter-laboratory comparison. Ann Bot 82:17–26
Doležel J, Greilhuber J, Suda J (2007) Estimation of nuclear DNA content in plants using flow cytometry. Nat Protoc 2:2233–2244
Dzialuk A, Chybicki I, Welc M, Sliwinska E, Burczyk J (2007) Presence of triploids among oak species. Ann Bot 99:959–964
Enke N, Fuchs J, Gemeinholzer B (2011) Shrinking genomes? Evidence from genome size variation in Crepis (Compositae). Plant Biol 13:185–193
Galbraith DW (2009) Simultaneous flow cytometric quantification of plant nuclear DNA contents over the full range of described angiosperm 2C values. Cytometry Part A 75A:692–698
Galbraith DW, Harkins KR, Maddox JM, Ayres NM, Sharma DP, Firoozabady E (1983) Rapid flow cytometric analysis of the cell-cycle in intact plant tissues. Science 220:1049–1051
Gmitter FG Jr, Chen C, Machado MA, de Souza AA, Ollitrault P, Froehlicher Y, Shimizu T (2012) Citrus genomics. Tree Genet Genomes 8:611–626
Govaerts R, Frodin DG (1998) World checklist and bibliography of Fagales (Betulaceae, Corylaceae, Fagacaea and Ticodendraceae). Royal Botanic Gardens, Kew
Grattapaglia D, Vaillancourt RE, Shepherd M, Thumma BR, Foley W, Külheim C, Potts BM, Myburg AA (2012) Progress in Myrtaceae genetics and genomics: Eucalyptus as the pivotal genus. Tree Genet Genomes 8:463–508
Gregory TR (2005) The C-value enigma in plants and animals: a review of parallels and an appeal for partnership. Ann Bot 95:133–146
Grime JP, Mowforth MA (1982) Variation in genome size—an ecological interpretation. Nature 299:151–153
Hall SE, Dvorak WS, Johnston JS, Price HJ, Williams CG (2000) Flow cytometric analysis of DNA content for tropical and temperate New World pines. Ann Bot 86:1081–1086
Hiremath SC, Nagasampige MH (2004) Genome size variaiton and evolution in some species of Dalbergia Linn.f. (Fabaceae). Caryologia 57(4):367–372
Hudson CJ, Kullan ARK, Freeman JS et al (2011) High synteny and colinearity among Eucalyptus genomes revealed by high-density comparative genetic mapping. Tree Genet Genomes 8:339–352. doi:10.1007/s11295-011-0444-9
Jedrzejczyk I, Sliwinska E (2010) Leaves and seeds as materials for flow cytometric estimation of the genome size of 11 Rosaceae woody species containing DNA-staining inhibitors. J Bot. doi:10.1155/2010/930895
Kremer A, Casasoli M, Berreneche T, Bodenes C, Sisco P, Kubisiak TL, Scalfi M, Leonardi S, Bakker E, Buiteveld J, Romero-Severson J, Arumuganathan K, Derory J, Scotti-Saintagne C, Roussel G, Bertocchi ME, Lexer C, Porth I, Hebard F, Clark C, Carlson J, Plomion C, Koelewijn HP, Villani F (2007) Fagaceae trees. In: Kole C (ed) Forest Trees. Springer, Berlin, pp 161–187
Kremer A, Abbott AG, Carlson JE, Manos PS, Plomion C, Sisco P, Staton ME, Ueno S, Vendramin GG (2012) Genomics of Fagaceae. Tree Genet Genomes 8:583–610
Leitch IJ, Chase MW, Bennett MD (1998) Phylogenetic analysis of DNA C-values provides evidence for a small ancestral genome size in flowering plants. Ann Bot 82:85–94
Levin DA, Funderburg SW (1979) Genome size in angiosperms: temperate versus tropical species. Am Nat 114:784–795
Lotsy JP (1925) Species or linneon. Genetica 7:487–506. doi:10.1007/BF01676287
Lotsy JP (1931) On the species of the taxonomist in its relation to evolution. Genetica 13:1–16. doi:10.1007/BF01725037
Loureiro JO, Rodriguez E, DOLE ELJ, Santos C (2006) Flow cytometric and microscopic analysis of the effect of tannic acid on plant nuclei and estimation of DNA content. Ann Bot 98:515–527
Manos PS, Stanford AM (2001) The historical biogeography of Fagaceae: tracking the tertiary history of temperate and subtropical forests of the Northern Hemisphere. Int J Plant Sci 162:77–93
Manos PS, Doyle JJ, Nixon KC (1999) Phylogeny, biogeography, and processes of molecular differentiation in Quercus subgenus Quercus (Fagaceae). Mol Phylogenet Evol 12:333–349
Manos PS, Zhou ZK, Cannon CH (2001) Systematics of Fagaceae: Phylogenetic tests of reproductive trait evolution. Int J Plant Sci 162:1361–1379
Manos PS, Cannon CH, Oh S-H (2008) Phylogenetic relationships and taxonomic status of the paleoendemic Fagaceae of western North America: recognition of a new genus, Notholithocarpus. Madrono 55:181–190
Mittelbach GG, Schemske DW, Cornell HV, Allen AP, Brown JM, Bush MB, Harrison SP, Hurlbert AH, Knowlton N, Lessios HA (2007) Evolution and the latitudinal diversity gradient: speciation, extinction and biogeography. Ecol Lett 10:315–331
Neale DB, Kremer A (2011) Forest tree genomics: growing resources and applications. Nat Rev Genet 12:111–122. doi:10.1038/nrg2931
Nixon KC (1993) Infrageneric classification of Quercus (Fagaceae) and typification of sectional names. Ann For Sci 50:25–34
Oh SH, Manos PS (2008) Molecular phylogenetics and cupule evolution in Fagaceae as inferred from nuclear CRABS CLAW sequences. Taxon 57:434–451
Ohri D (2005) Climate and growth form: the consequences for genome size in plants. Plant Biol 7:449–458
Oliver MJ, Petrov D, Ackerly D et al (2007) The mode and tempo of genome size evolution in eukaryotes. Genome Res 17:594–601. doi:10.1101/gr.6096207
Pellicer J, Fay MF, Leitch IJ (2010) The largest eukaryotic genome of them all? Bot J Linn Soc 164:10–15
Petit RJ, Hampe A (2006) Some evolutionary consequences of being a tree. Annu Rev Ecol Evol Syst 37:187–214
Price HJ, Hodnett G, Johnston JS (2000) Sunflower (Helianthus annuus) leaves contain compounds that reduce nuclear propidium iodide fluorescence. Ann Bot 86:929–934
Soepadmo E (1972) Fagaceae. In: Soepadmo E (ed) Flora Malesiana, vol 7. vol. 1. Noordhoff International, Leyden, pp 265–403
Soltis DE, Soltis PS (1999) Polyploidy: recurrent formation and genome evolution. Trends Ecol Evol 14:348–352
Soltis DE, Soltis PS, Bennett MD, Leitch IJ (2003) Evolution of genome size in the angiosperms. Am J Bot 90:1596–1603
Soltis DE, Albert VA, Leebens-Mack J, Bell CD, Paterson AH, Zheng C, Sankoff D, de Pamphilis CW, Wall PK, Soltis PS (2009) Polyploidy and angiosperm diversification. Am J Bot 96:336–348
Stebbins GL (1938) Cytological characteristics associated with the different growth habits in the dicotyledons. Am J Bot 25:189. doi:10.2307/2436589
Wendel JF, Cronn RC, Spencer Johnston J, James Price H (2002) Feast and famine in plant genomes. Genetica 115:37–47
Wood TE, Takebayashi N, Barker MS, Mayrose I, Greenspoon PB, Rieseberg LH (2009) The frequency of polyploid speciation in vascular plants. Proc Natl Acad Sci 106:13875–13879
Zoldos V, Papes D, Brown SC, Panaud O, Siljak-Yakovlev S (1998) Genome size and base composition of seven Quercus species: Inter- and intra-population variation. Genome 41:162–168
Acknowledgements
We thank J. Doležel for providing the seeds of internal standards; Y-H Tan and the XTBG Seed Bank for acorn collection; I. Leith, J. E. Carlson, Z-K Zhou, and G. M. Lambert for beneficial communication; and 2009 Yunnan High-End Talent of Yunnan Regional Government (No. 09SK051) and the National Basic Research Program of China (973 Program, No. 08GK014B01) for funding support.
Data Archiving Statement
The data in this analysis consists of flow cytometry measurements of nuclear DNA content for 171 individuals from 47 tropical Chinese species, including three separate genera and the tropical subgenus Cyclobalanopsis within the genus Quercus. This data will be deposited at the Plant DNA C-values Database (http://data.kew.org/cvalues/) hosted by the Royal Botanic Gardens, Kew, UK.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Kremer
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(JPEG 575 kb)
Rights and permissions
About this article
Cite this article
Chen, SC., Cannon, C.H., Kua, CS. et al. Genome size variation in the Fagaceae and its implications for trees. Tree Genetics & Genomes 10, 977–988 (2014). https://doi.org/10.1007/s11295-014-0736-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11295-014-0736-y