Abstract
One of the most dynamically developing sectors of green biotechnology is molecular farming using transgenic plants as natural bioreactors for the large scale production of recombinant proteins with biopharmaceutical and therapeutic values. Such properties are characteristic of certain proteins of bacterial origin, including staphylokinase. For many years, work has been carried out on the use of this protein in thrombolytic therapy. In this study, transgenic Solanum tuberosum plants expressing a CaMV::sak-mgpf-gusA gene fusion, were obtained. AGL1 A. tumefaciens strain was used in the process of transformation. The presence of the staphylokinase gene was confirmed by PCR in 22.5% of the investigated plants. The expression of the fusion transgene was detected using the β-glucuronidase activity assay in 32 putative transgenic plants. Furthermore, on the basis of the GUS histochemical reaction, the transgene expression pattern had a strong, constitutive character in seven of the transformants. The polyacrylamide gel electrophoresis of a protein extract from the SAK/PCR-positive plants, revealed the presence of a119 kDa protein that corresponds to that of the fusion protein SAK-mGFP-GUSA. Western blot analysis, using an antibody against staphylokinase, showed the presence of the staphylokinase domain in the 119 kDa protein in six analyzed transformants. However, the enzymatic test revealed amidolytic activity characteristic of staphylokinase in the protein extract of only one plant. This is the first report on a Solanum tuberosum plant producing a recombinant staphylokinase protein, a plasminogen activator of bacterial origin.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The possibility of using plants as a production system for the proteins of pharmaceutical value was demonstrated in the late 1990s, when the successful expression of a fusion protein of the human growth hormone and a functional antibody was accomplished (Fischer et al. 2004). These remarkable achievements showed the significance of plants for the production of mammalian conjugated proteins. Moreover, with molecular farming, it is possible to obtain considerable amounts of an active and pharmacologically safe product using low financial outlays. Currently, technologies based on plant expression systems are used for the production of recombinant antibodies (rAbs), enzymes, hormones, interleukins, and vaccines (Tiwari et al. 2009). To this day, numerous transformation procedures for various plant species have been elaborated (Schillberg et al. 2005). Many of these plant species have been used for the production of biopharmaceuticals for clinical tests (Decker and Reski 2007). In recent years, a limited number of veterinary biopharmaceuticals and vaccines produced in plant cell cultures were issued a marketing permission (Twyman et al. 2005; Walsh 2006).
Thrombolytic therapy has become one of the routine ways of treating acute myocarditis. As a result, efficiency, specificity and tolerance of thrombolytic drugs have been improved in the recent years, and this improvement was confirmed through pharmacological and clinical studies. A common characteristic of new thrombolytic factors is their durability in plasma. The role of fibrinolytic factors, which are also called plasminogen activators, consist of transforming a zymogen (plasminogen) into its enzymatically active form, plasmin (Collen and Lijnen 1991). Plasmin, in turn, digests fibrinogen deposits, which results in dissolving a clot, consequently restoring the patency of a vessel. As a result of plasmin activity, the restoration of proper blood circulation conditions occurs.
Staphylokinase (SAK) is a protein produced and secreted by some strains of Staphylococcus aureus and S. epidermidis, S. lentus, S. sciuri, S. lugdunesis, S. xylosus, and S. hominis (Bokarewa et al. 2006). Initially, staphylokinase was obtained from natural biological material, i.e. S. aureus cultures. Presently, many research teams use various expression systems to produce this protein, e.g. E. coli, P. mirabilis, P. pastoris, B. subtilis and S. hygroscopicus (Gehmlich et al. 1997; Miele et al. 1999; Szarka et al. 1999; Rajamohan and Dikshit 2000). The mechanism of action of staphylokinase is currently well known and it has been precisely characterized. It is known that as a plasminogen activator, it displays a highly specific activity towards fibrin, and it is considered to be a promising thrombolytic factor, specifically stimulating the thrombolysis of both thrombocyte- and erythrocyte-rich clots (Szarka et al. 1999). Like streptokinase (STK), SAK displays no proteolytic activity; instead, it is a cofactor that forms a non-active, stoichiometric 1:1 complex with plasminogen. This complex requires plasminogen conversion into plasmin in order to exhibit its active sites, as well as the amino-terminal treatment of SAK, which induces the potential of plasminogen activator (Rajamohan and Dikshit 2000).
The aim of this study was to obtain prototype transgenic potato plants that produce recombinant staphylokinase. In the study, the plant expression cassette used was engineered using a binary plasmid CAMBIA 1304, harboring the selective marker CaMV::htp and the transgene encoding the fusion protein STAF-GFP-GUS (Wiktorek-Smagur et al. 2011). Although the production of recombinant staphylokinase was confirmed by Western blot analysis in only six independent transformants, and the presence of the protein cofactor activity was confirmed by the amidolytic assay in only one of them, this is the first report on a crop plant producing a recombinant staphylokinase protein.
Materials and methods
Plant material
In the study, Solanum tuberosum cv. Desiree was used. Potato plants were grown and micropropagated in vitro on a standard Murashige and Skoog (1962) medium, according to the protocol obtained courtesy of Professor J. Hennig from the Institute of Biochemistry and Biophysics of the Polish Academy of Sciences in Warsaw, Poland.
Bacterial strains
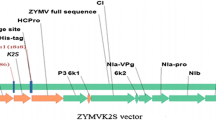
AGL1 strain of Agrobacterium tumefaciens with a recombinant plasmid pCAMBIA 1304 (Centre for the Application of Molecular Biology to International Agriculture, Canberra, Australia) was used for the transformation process. The T-DNA region of the original plasmid consisted of a plant-selective hygromycin resistance gene and two reporter markers in the form of a gene fusion mgfp5-gusA-His6 (http://www.cambia.org.au; Fig. 1—the expression cassette outline). The recombinant binary plasmid carried the gene fusion of staphylokinase and two reporter genes (mgfp5 and gusA) driven by the CaMV 35S promoter.
Outline of the T-DNA construct of pCAMBIA 1304sak plasmid indicating the location of sak sequence insert. The amplified fragment of the staphylokinase gene (accession number M57455—GenBank) was cloned into a binary plasmid pCAMBIA 1304, which was verified by PCR and NcoI and BglII restriction analysis. The staphylokinase gene was placed under the control of the 35S promoter of CaMV
1304sak plasmid construction
The staphylokinase gene used in the study was obtained from pTrxFusSTA (Fig. 2). With the use of this plasmid as a template, the amplification of the sak sequence was performed using the following primers:
-
8—5′ GAC CAT GGG ATG ATC AAA AGA 3′ and
-
9—5′ GCA CTA GTT TTC TTT TCT ATA 3′.
The staphylokinase gene used in the study was cloned into a pCAMBIA 1304 plasmid between the sequences recognized by the restriction enzymes NcoI and BglII.
Introduction of the 1304 plasmid with the sak gene into Agrobacterium tumefaciens
The recombinant binary plasmid 1304sak was mobilized into Agrobacterium tumefaciens using the triparental mating procedure (Wise et al. 2006).
Plant transformation and regeneration
Transgenic potato plants were obtained by Agrobacterium-mediated transformation via co-cultivation with leaf explants derived from 6-week-old potato plants grown in vitro. One hundred fifty μL aliquots of a bacterial suspension were spread thinly on the surface of MS medium. On the thus prepared media, leaf blade disc explants were placed in such a way that the upper side of the blades adhered to the medium. Petri dishes containing explants (not sealed with parafilm) were left for 48 h in the dark at a room temperature. During this incubation period, the growth of Agrobacterium tumefaciens was observed and the explants became infected. After two days of co-cultivation with the bacteria on the antibiotic-free medium, explants were transferred to a callus inducing medium (MCI; MS + BAP 0.1 mg/mL, NAA 5 mg/L), supplemented with relevant antibiotics: 25 mg/L hygromycin B and 500 mg/L cefotaxime. The explants were grown for 14 d under 16/8 h photoperiod, at 26 °C. Explants producing callus tissues were placed on shoot inducing medium (GR2; MS + 0.02 mg/l GA3, 0.02 mg/l NAA) which contained the above mentioned antibiotics in the same concentrations. The cultures were subcultured for several passages on a fresh medium at an interval of 2 weeks. After 7–9 d, the formation of the first small shoots was successfully observed. They were then transferred to culture tubes onto an MS selection medium supplemented with antibiotics.
Histochemical analysis of β-glucuronidase (GUS) activity
The histochemical analysis of the GUS activity was performed according to Kononowicz et al. (1992). 6 to 12 μm thick sections were mounted on microscope slides. Microscopic analyses (paraffin sections of leaf blade fragments) were performed using the light microscope Olympus BX60. The images of the stained plants were visualized and captured using a Nikon SMZ 800 stereoscopic microscope and a computer image analysis program (Soft Imaging System GmbH, Germany).
Genomic DNA isolation
Isolation of DNA from S. tuberosum leaves and microtubers was performed according to the modified Dellaporta et al. (1983) method.
DNA amplification with the use of the PCR technique
The integration of transgenes into the S. tuberosum genome was confirmed by PCR using genomic DNA as a template. DNA amplification with PCR was performed using the Sambrook et al. (1989) method, with slight modifications. PCR was performed with a Biometra thermocycler (Germany). Primers STAF 8 (5′ GAC CAT GGG ATG ATC AAA AGA 3′) and STAF 9 (5′ GCA CTA GTT TTC TTT TCT ATA 3′) were designed to amplify the sak region of the plasmid, encoding staphylokinase. The amplifications were carried out as follows: 95 °C for 4 min, followed by 30 cycles at 56 °C for 10 s, 72 °C for 30 s and 94 °C for 10 s. The final elongation step was carried out at 72 °C for 4 min.
Protein isolation
Protein isolation was performed according to the Kaźmierczak and Kaźmierczak (2003) method.
Western blot analysis and protein staining with Coomassie Brilliant Blue
Western blot analysis was performed according to the method described by Sambrook et al. (1989). Protein separation by SDS–polyacrylamide gel electrophoresis was performed at 120 V. The analysis of protein electrophoretograms stained for 1 h with Coomassie Brilliant Blue was carried out using the Image Master VDS Software computer program (Pharmacia Biotech). The proteins were transferred to a nitrocellulose membrane for immunoblotting. The primary antibodies (mouse polyclonal anti-staphylokinase, kindly provided by Professor J. Szemraj, Lodz Medical University, Poland), were used at 1:5,000 dilutions and the secondary antibodies (alkaline phosphatase-conjugated anti-mouse, Sigma-Aldrich, USA) were used at 1:2,000 dilutions. Western blot analyses were performed using the Bio-Rad Western blotting apparatus (USA).
Analysis of the amidolytic activity of staphylokinase in the protein extracts
The measurement of the amidolytic activity of the protein extract was performed by determining the absorbance difference between the obtained pNA and that of the original substrate. The plasminogen activation assay involves a sequential reaction as follows:
where PA (plasminogen activators); Plg (plasminogen), Plm (plasmin), pNA (p-nitroaniline) and H–D-Val–Leu–Lys–OH are products of the S-2251 hydrolysis (D-Val–Leu–Lys-p-nitroanilide, S2251; Chromogenix, USA). In order to determine the amidolytic activity, 40 μL of protein extract from S. tuberosum was added to 5 mM sodium phosphate buffer (pH 7.4) containing 0.15 M NaCl and 1.1 μM of plasminogen (Plg, isolated from human plasma). The reference sample did not contain plasminogen. After incubation for 30 min at 37 °C, the reaction was interrupted by adding S2251 (0.5 mM H–D-Val–Leu–Lys-pNA in 50 mM Hepes–NaOH, pH 7.4). The absorbance was measured at 405 nm. The results were read from a calibrated diagram prepared with the use of p-nitroaniline (pNa) standard solutions.
Results
Regeneration of in vitro plants
In the present study, following the transformation using A. tumefaciens, regenerated S. tuberosum cv. Desiree plants were obtained under selective pressure (Fig. 3h). Four weeks later, callus formation was observed (Fig. 3a, b), and approximately 2.5 months after the start of the experiment, the formation of new shoots took place (Fig. 3c–g). In the case of the negative control, progressive chlorosis of explants was detected which was a consequence of the toxic activity of antibiotics (cefotaxime and hygromycin B). Leaf explants, which consisted of the positive control, showed a morphogenetic response in a relatively short time (4 weeks following agroinfection) as compared to the vector control (10 weeks). In general, in regenerated plants, both in the case of the positive and vector controls, no morphological abnormalities, or growth and development disorders were observed (Fig. 4). However, single cases were recorded in which the leaves were slightly smaller and the plant more bushy (data not shown). All the obtained plantlets were able to regenerate under selective pressure conditions (500 mg/L cefotaxime and 25 mg/L hygromycin B).
PCR analysis
Seventy-one plants (putative transgenics) regenerated under the conditions of selective pressure, and were subjected to PCR analysis. Samples of twelve microtubers were also subjected to PCR analysis. The electrophoresis of PCR products of the examined plants, which used genomic DNA as template, showed single bands (about 500 bp size) corresponding to the staphylokinase-coding sequence in 16 plants (Fig. 5b). Among the 12 microtubers tested, all showed a single band of ~500 bp size corresponding to the staphylokinase-coding sequence (Fig. 5a).
Detection of the coding sequence (sak) in transgenic S. tuberosum cv. Desiree plants. a Example of PCR analysis of the binary vector 1304sak to detect the presence of the sak gene in transgenic lines of S. tuberosum cv. Desiree (genomic DNA as a template, isolated from microtubers) (lane 1 positive control recombinant staphylokinase (SAK+) from E. coli transformants; lanes 2–4 independent transgenic lines; lane 5 a wild-type plant; lane 6 negative control; lane 7 the molecular weight markers (M); b example of PCR analysis of the binary vector 1304sak to detect the presence of the sak gene in transgenic lines of S. tuberosum cv. Desiree (genomic DNA as a template isolated, from leaves) (lane 1 the molecular weight markers (M); lane 2 a wild-type plant; lanes 3–9 independent transgenic lines; lane 10 positive control recombinant staphylokinase (SAK+) from E. coli transformants
Histochemical GUS activity reaction
The histochemical analysis of GUS activity confirmed gusA gene expression in the examined leaves and microtubers of S. tuberosum (respectively Fig. 6b, d). The gusA gene expression patterns in the examined leaves varied in terms of color distribution and intensity, whereas in examined microtubers it was evenly distributed (Fig. 6d). In wild plant tissues, which were the negative control, no GUS activity was observed (Fig. 6a, c). Macroscopic observations were confirmed by the microscopic analysis (paraffin sections of leaf blade fragments) performed using the light microscope (Fig. 6e). In the analyzed tissue sections, the β-glucuronidase activity product appearing as a blue color precipitate was found both in the mesophyll and the epidermis. Of note, all putative transgenic plants that we obtained, spontaneously produced microtubers, at the count of one per plant, all of which were found to be GUS positive as evidenced by histochemical assay.
Histochemical demonstration of GUS enzyme activity in transgenic S. tuberosum cv. Desiree plants. Putative transgenic plants were assayed histochemically for GUS activity as described in “Materials and methods”. a Leaf from the non-transformed plant; b GUS activity was detected in the leaves of putative transgenic plants. c Microtuber from non-transformed plant; d GUS activity was detected in the microtubers of putative transgenic plants; e cross-section of leaf fragments from GUS-positive putative transgenic plants. The β-glucuronidase activity product was found both in mesophyll and epidermis
Western blot analysis
Protein extracts from leaf fragments of selected transgenic and control plants were subjected to western blot analysis and the amidolytic activity assay. Six selected S. tuberosum plants regenerated under the conditions of selective pressure and, showing GUS activity, were examined using the SDS–PAGE and the western blot analysis (Fig. 7a, b). Previously, the presence of the staphylokinase coding sequence was revealed in all of these plants using the PCR method. In two cases, western immunoblot analysis revealed a cross-reaction of anti-staphylokinase antibodies with 119 kDa protein present in the extracts from leaf fragments (Fig. 7b). It should be emphasized that the plants containing the staphylokinase domain in the protein whose molecular weight was 119 kDa (corresponding to the molecular weight of the fusion protein, the product of CaMV::sak-mgfp-gusA transgene), also showed a high GUS activity.
Protein profiles and Western blot analysis of CaMV::sak-mgfp-gusA in transgenic S. tuberosum cv. Desiree plants. a SDS–PAGE gels visualized by coomassie blue staining. The protein molecular weight markers (MW); lanes 1, 3, 4 independent transgenic lines; lane 2 a wild-type plant. b The identity of SAK was established using the Western immunoblots. Three independent transgenic lines are shown (lanes 1,3 and 4); lane 2 a wild-type plant
Analysis of amidolytic activity in protein extracts
Amidolytic activity was detected in the protein extract of only one transgenic plant (Fig. 8). No such activity was observed in the control plants or in other protein extract samples.
Discussion
Improving S. tuberosum cultivars using traditional breeding methods is difficult due to the tetraploidity and heterozygosity of this species as well as to its frequent fertility disorders. On the other hand, Solanum tuberosum is a member of Solanaceae family, whose representatives easily regenerate in tissue cultures and are susceptible to the A. tumefaciens-mediated transformation method. The relatively fast growth of potato plants and their susceptibility to genetic transformation and microtuber generation make this plant an attractive subject for recombinant protein production, including vaccines. For these reasons, S. tuberosum is a species widely used in molecular farming studies of recombinant proteins, and particularly in terms of vaccine production. For different cultivars of potatoes, many regeneration and transformation procedures have been reported. In this study, leaf blade fragments were used for agroinfection. The explants came from the youngest (1–4) leaves of donor plants whose young tissues usually provide the best morphogenetic response. Macroscopic analysis showed that, in the positive control, young shoots appeared with a frequency of about 5–6 per explant, while in the vector control, 2–3 shoots. After the co-cultivation of the explants with A. tumefaciens, it was necessary to immediately eliminate bacteria from the culture. Effective antibiotic concentrations are usually not harmful for plant tissues, but are found to affect the in vitro morphogenetic processes. For instance, cefotaxime at a concentration of 500 mg/L brings about a decreased regeneration in such species as A. thaliana, D. carota, M. domestica, P. alba, T. avestivum, Malus sp., P. glauca, P. communis and S. tuberosum (Nauerby et al. 1997). In addition, the negative effect of cefotaxime in doses of 250–500 mg/L, on the induction, growth and differentiation of callus tissue from leaf explants, was demonstrated by Xiangqian et al. (2002) in their studies on Rosa hybrida. In this study, cefotaxime was used at a high concentration (500 mg/L), and this fact may explain the low frequency and slow regeneration of plants in the vector control.
In order to eliminate or weaken the cells that did not undergo transformation, a selective antibiotic is added to the medium. Following the recommendations of the binary vector producer, CAMBIA (http://www.cambia.org.au/daisy/bioforge_legacy/3726.html), hygromycin B at the concentration of 25 mg/L was used. The most visible effect of the antibiotic action on the untransformed tissues was their progressive chlorosis and, consequently, the explant’s death. When the action of the selective agent is prolonged, the growth, proliferation, and rooting of the shoots is significantly inhibited. As expected, these effects were observed in the negative control. Of the explants subjected to the transformation procedure under selective pressure, 71 healthy plants were obtained, each with a well developed radical system and a normal phenotype. Twelve plants spontaneously produced microtubers, at the count of one per plant. We did not purposefully induce microtubers. This result is rather important since tissue cultures can generate both epigenetic and hereditary changes, in the form of somaclonal variations and mutations. It is also well documented that somaclonal variation may affect transgene expression. The expression level of foreign genes can however also have a strong negative effect on the plant phenotype, such as sterility and dwarfness (compare transgenic rice plants expressing lectin; Bhat and Srinivasan 2002).
In the present study, a sak-mgfp-gusA gene fusion under the control of the CaMV promoter was used for the transformation of S. tuberosum. All 71 plants, obtained under selective pressure, including 12 microtubers, were tested for GUS activity. The GUS expression was detected in 32 plants, which represents 45% of the putative transgenics, and in all cases of microtubers. The transgene expression level assessed on the basis of staining intensity was diverse: in seven putative GUS-positive transformants, representing 9.8% of all putative transformants, tissue staining was very strong; in the remaining 64, a moderate or very weak color intensity was displayed. A similar effect was observed in the case of microtubers. In some cases of the so-called false positive results, the detected signal is thought to have been caused by an endophytic bacteria showing β-glucuronidase expression. This kind of activity can result in masking the activity coming from the reporter gene introduced in the plant, especially when its expression is on the threshold of detection. Taking into account the high effectiveness of the method of eliminating A. tumefaciens from the plant material, and the spatial pattern of the chromatic reaction, we believe that the low level of β-glucuronidase in the plants constituting 90.1% of the putative transformants population was due to the effect of the low transgene expression or to the intrinsic beta-glucuronidase activity, rather than to activity of bacterial origin.
PCR analysis of the 12 microtubers showed the presence of the transgene in all cases, and the positive amplification result was also correlated with the positive result of the histochemical GUS assay. However, PCR analysis of the 71 plants showed the presence of the transgene in only 22.5% of transformants. The positive amplification result did not however correlate with the positive result of the histochemical GUS assay in all cases. Similarly, in the study carried out by Hiei and Kubo (1997), not all of the rice plants obtained on selective medium after A. tumefaciens transformation displayed a gusA gene expression. Notwithstanding this, Southern blot analysis showed that the plants did have an integrated transgene, in one or several copies. Emani et al. (2002) observed a similar phenomenon in their study on African oat. The authors explained this as an effect of gene silencing.
Another reason for low gene expression of bacterial transgenes in plants can be the presence of cryptic amber codons, false signals of polyadenylation or splicing. This was reported by Haffani et al. (2000) who found that the cause of a very low level of cry3Cal gene expression in potato is premature transcript adenylation brought about by the presence, within the open reading frame, of sequences recognized by the eukaryotic transcription apparatus as a polyadenylation signal.
The results presented in this study pertain to the integration of the fusion gene CaMV::sak-mgfp-gusA to the Solanum tuberosum plant genome. In the case of two out of six examined transformants, western blot analysis allowed the observation of staphylokinase via immunochemical detection and identification. In terms of the amidolytic activity of the protein extract, the performed biochemical tests confirmed it in only one plant. These results could indicate, on one hand, a very low production of staphylokinase, and on the other, low amidolytic activity of the fusion protein. It is probable that this protein occurs in the cells of other transformants, but in such low quantity that it cannot be detected by western blot analysis. A similar phenomenon was observed by Wiktorek-Smagur et al. (2011) in reference to staphylokinase. In their study on Solanum tuberosum, Hassairi et al. (1998) observed a similar effect in reference to the coat protein LMV. Low content of staphylokinase in protein extracts can also explain its low amidolytic activity, or even the complete lack thereof.
Biological activity of proteins and their functionality is conditioned by a proper posttranslational folding. The product of the transgene used in the study is a complex fusion protein with the molecular weight of 119 kDa. Therefore, another reason for the low amidolytic activity of the transgene’s protein product may be its improper posttranslational folding, during which the staphylokinase domain could have remained inappropriately exposed since it may be covered by reporter protein domains (Zhang et al. 2002).
Transgene expression can also be limited by the large sizes of the coding sequence. In the present study, the transgene used for the transformation of S. tuberosum was of a noteworthy size, which is connected with the presence of both gusA and mgfp coding sequences. The length of the whole gene construct amounted to 3,057 bp. The elimination of reporter genes and consequently, the reduction of the transgene size, should undoubtedly favor both its proper integration with the plant genome and the increased production of recombinant staphylokinase.
The pCAMBIA plasmids enable the construction of translational gene fusions. The fusion of staphylokinase with reporter genes in the present study, necessary due to the need to monitor transgene expression in successive stages of the transformation and regeneration processes, may have had an important influence on the gene expression level, the protein content in cells, and the amidolytic activity of staphylokinase. It could also be the reason for the low or even completely reduced β-glucuronidase quantity detected in plants in which the presence of the transgene was confirmed by PCR. Currently, research is being carried out on recombinant fusion proteins, containing staphylokinase coding sequence as one of the components. As it follows from literature data, hybrid constructions based on staphylokinase are widely used (Chen et al. 2007; Szemraj et al. 2005, 2007; Chiou et al. 2007).
In conclusion, we hereby report, presumably for the first time, the possibility of obtaining transgenic potato plants which produce recombinant staphylokinase, an effective thrombolytic agent of bacterial origin.
This research is currently being continued in our laboratory in order to obtain successive vegetative generations of plants from the transgenic tubers, and to produce transgenic plants utilizing new expressing cassettes, consisting of the staphylokinase coding sequence, under control of patatin (B33) and other promoters.
Abbreviations
- AGL1:
-
Agrobacterium tumefaciens strain
- BAP:
-
6-Benzylaminopurine
- Gfp :
-
Gen coding green fluorescent protein
- GR2:
-
Shoot inducing medium
- gusA :
-
β-Glucuronidase
- hPm:
-
Human plasmin
- htp :
-
Neomycin phosphotransferase
- MCI:
-
Callus inducing medium
- Plg:
-
Plasminogen
- Plm:
-
Plasmin
- pNA:
-
p-Nitroaniline
- r-SAK:
-
Recombinant staphylokinase
- S-2251:
-
D-Val–Leu–Lys-p-nitroanilide
- SAK:
-
Staphylokinase
References
Bhat SR, Srinivasan S (2002) Molecular and genetic analyses of transgenic plants: considerations and approaches. Plant Sci 163:673–681
Bokarewa MI, Jin T, Tarkowski A (2006) Staphylococcus aureus: staphylokinase. Int J Biochem Cell Biol 38:504–509
Chen H, Mo W, Zhang Y, Su H, Ma J, Yao R, Zhang S, Ge J, Song H (2007) Functional properties of a novel mutant of staphylokinase with platelet-targeted fibrinolysis and antiplatelet aggregation activities. Eur J Pharmacol 566:137–144
Chiou JF, Woon MD, Cheng SN, Hsu CH, Cherng SC, Hsieh FK, Lin SM, Shiau CY (2007) Staphylokinase-annexin XI cimera exhibited efficient in vitro thrombolytic activities. Biosci Biotechnol Biochem 71:1122–1129
Collen D, Lijnen HR (1991) Basic and clinical aspects of fibrynolysis and thrombolysis. Blood 78:3114–3124
Decker EL, Reski R (2007) Moss bioreactors improved biopharmaceuticals. Curr Opin Biotechnol 18:393–398
Dellaporta AL, Wood J, Hicks JB (1983) Plant DNA minipreparations. Version II. Plant Mol Biol Rep 1:19–71
Emani C, Sunilkumar G, Rathore KS (2002) Transgene silencing and reactivation in sorghum. Plant Sci 162:181–192
Fischer R, Stoger E, Schillberg S, Christou P, Twyman RM (2004) Plant-based production of biopharmaceuticals. Curr Opin Plant Biotechnol 7:152–158
Gehmlich I, Pohl HD, Knorre WA (1997) Laboratory-scale permeabilization of Escherichia coli cells for recovery of a small recombinant protein–staphylokinase. Bioprocess Eng 17:35–38
Haffani YZ, Overney S, Yelle S, Bellemare G, Belzile FJ (2000) Premature polyadenylation contributes to the poor expression of the Bacillus thuringiensis cry3Ca1 gene in transgenic potato plants. Mol Gen Genet 264:82–88
Hassairi A, Masmoudi K, Albouy J, Robaglia C, Jullien M, Ellouz R (1998) Transformation of two potato cultivars “Spunta” and “Claustar” (Solanum tuberosum) with lettuce mosaic virus coat protein gene and heterologous immunity to potato virus Y. Plant Sci 136:31–42
Hiei Y, Kubo T (1997) Transformation of rice mediated by Agrobacterium tumefaciens. Plant Mol Biol 35:205–218
Kaźmierczak A, Kaźmierczak J (2003) Application of capillary electrophoresis for DNA-protein binding tests. Acta Physiol Plant 25:13–17
Kononowicz AK, Nelson DE, Singh NK, Hasegawa PM, Bressan RA (1992) Regulation of the osmotin gene promoter. Plant Cell 4:513–524
Miele RG, Prorok M, Costa VA, Castellino J (1999) Glycosylation of asparagine-28 recombinant staphylokinase with high—mannose-type oligosaccharides results in a protein with highly attenuated plasminogen activator activity. J Biol Chem 274:7769–7776
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15:473–497
Nauerby B, Billing K, Wyndaele R (1997) Influence of the antibiotic timentin on plant regeneration compared to carbenicillin and cefotaxime in concentrations suitable for elimination of Agrobacterium tumefaciens. Plant Sci 123:169–177
Rajamohan G, Dikshit KL (2000) Role of the N-terminal region of staphylokinase (SAK): evidence for the participation of the N-terminal region SAK in the enzyme–substrate complex formation. FEBS Lett 474:151–158
Sambrook J, Maniatis T, Fritsch EF (1989) Molecular cloning. CSH Laboratory Press, New York
Schillberg S, Twyman RM, Fischer R (2005) Opportunities for recombinant antigen and antibody expression in transgenic plants-technology assessment. Vaccine 23:1764–1769
Szarka SJ, Sihota EG, Habibi HR, Wong SL (1999) Staphylokinase as a plasminogen fusion activator component in recombinant fusion proteins. Appl Envir Microbiol 65:506–513
Szemraj J, Walkowiak B, Kawecka I, Janiszewska G, Buczko W, Bartkowiak J, Chabielska E (2005) A new recombinant thrombolytic and antithrombotic agent with higher fibrin affinity—a staphylokinase variant. I. In vitro study. J Thromb Haemos 3:2156–2165
Szemraj J, Stankiewicz A, Rozmyslowicz-Szermińska W, Mogielnicki A, Gromotowicz A, Buczko W, Oszajca K, Bartkowiak J, Chabielska E (2007) A new recombinant thrombolytic and antithrombotic agent with higher fibrin affinity—a staphylokinase variant. An in vivo study. Thromb Haemost 97:1037–1045
Tiwari S, Verma PC, Singh PK, Tuli R (2009) Plants as bioreactors for the production of vaccine antigens. Biotechnol Adv 27:449–467
Twyman RM, Schillberg S, Fischer R (2005) Transgenic plants in the biopharmaceuticals market. Expert Opin Emerg Drugs 10:185–218
Walsh G (2006) Biopharmaceutical benchmarks 2006. Nat Biotechnol 24:769–776
Wiktorek-Smagur A, Hnatuszko-Konka K, Gerszberg A, Łuchniak P, Kowalczyk T, Kononowicz AKK (2011) Expression of a staphylokinase, a thrombolytic agent in Arabidopsis thaliana. World J Microbiol Biotechnol 27:1341–1347
Wise AA, Lu Z, Binns AN (2006) Three methods for the introduction of foreign DNA into Agrobacterium. In: Wang K (ed) Methods in molecular biology. Humana Press Inc., Totowa, pp 43–54
Xiangqian L, Krasnyanski SF, Korban SS (2002) Optimization of the uidA gene transfer into somatic embryos of rose via Agrobacterium tumefaciens. Plant Physiol Biochem 40:453–459
Zhang X, Beuron F, Freemont PS (2002) Machinery of protein folding and unfolding. Curr Opin Struct Biol 12:231–238
Acknowledgments
We would like to thank Professor J. Szemraj (Department of Medical Biochemistry, Medical University of Lodz, Poland) for providing the pTrxFusSTA plasmid harboring staphylokinase coding sequence. This research was supported by grant No. 6 P04B 003 18 State Committee for Scientific Research, Poland.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Gerszberg, A., Wiktorek-Smagur, A., Hnatuszko-Konka, K. et al. Expression of recombinant staphylokinase, a fibrin-specific plasminogen activator of bacterial origin, in potato (Solanum tuberosum L.) plants. World J Microbiol Biotechnol 28, 1115–1123 (2012). https://doi.org/10.1007/s11274-011-0912-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11274-011-0912-2